
More microRNAs as biomarkers and hope for precision medicine in kidney diseases
microRNAs (miRNAs/miRs) are very short (20–25 nucleotides) single stranded noncoding RNAs which have the complementary to the 3' untranslated regions (3' UTRs) of target messenger RNAs (mRNAs) and induce the degradation of target RNAs and inhibit the protein translation (1-3). Those tiny RNAs are playing critical roles in physiological, developmental and pathological conditions and in variety of the human diseases and can be useful as biomarkers for human diseases because of their stable existence in body fluids such as blood and urine (4,5). Recent advances in the sensitive and quantitative detection of miRNAs in human biofluids are making miRNAs as promising noninvasive biomarkers for human diseases (6,7). Functional relevance of miRNAs in kidney diseases and their usefulness as biomarkers have been suggested, too (8,9). Comprehensive profiles of miRNAs in urine, urinary sediment and serum from patients in specific stages of DN, fibrosis, renal function decline [glomerular filtration rate (GFR)], albuminuria or rapid progression to end stage renal disease (ESRD) have been reported (10-16). Precise diagnosis at the early stage of kidney diseases is expected to provide the effective prevention of the diseases. A recent report identified numerous known and unknown miRNAs in urine as biomarkers for diabetic nephropathy (DN) and lupus nephritis (LN) (17). The authors profiled 2,402 urinary miRNAs in patients with DN or LN, and compared them with healthy controls. In DN, miR-2861, miR-1915-3p, and miR-4532 were significantly decreased and related to glomerular filtration rate and interstitial fibrosis/tubular atrophy. In LN patients, miR-3201 and miR-1273e were decreased and related to glomerular inflammation. Several members of miR-30 family were increased in both DN and LN and associated with glomerular filtration rate and proteinuria. New miRNAs identified in this study may provide specific noninvasive detection of DN and LN.
So far, numerous reports have been published for pursuing miRNAs as biomarkers for kidney diseases (8-16,18). It is still not easy to reach the clear conclusion because of some conflicting reports. However, molecular mechanisms might be different from patient to patient even in the same kidney disease. Big variations are observed in patient groups and even in healthy groups while the authors used healthy “patients” (17). Thus, another difficulty may be the availability of real healthy individuals. Deviations in patients may be caused by the stages in the disease progression. Some miRNAs may be high in the early stage but low in the late stage (13,14,18,19). Significant differences between DN and diabetes patients, and LN and systemic lupus erythematosus were detected in the same report (17). Therefore, simple comparison between healthy group and disease group may not be enough but the stage specificity of miRNAs might have to be studied.
Targeting certain miRNAs by chemically-modified antisense oligonucleotides has been successful to reduce specific miRNAs and inhibit fibrosis and hypertrophy in mouse models of DN and fibrosis (20-26). Treating patients with such miRNA inhibitors has been under development and evaluated in some clinical trials (27,28). Therefore, early detection of kidney diseases is benefit to prevent progression to renal failure and dialysis.
Combinations of newly identified and known miRNAs may segregate patients more precisely to several groups even in single kidney disease and provide personalized treatments targeting specific miRNAs (Figure 1). On the other hand, some patients may be categorized in the same group by miRNA profiling even if they had different diagnosis (DN, DL or others) and the same treatment targeting common particular miRNAs may be effective to treat the patients who have been diagnosed as different kidney diseases (Figure 1). Changes in early stage may be an indicator of the initiation of kidney diseases. Some patients may need treatments even before the obvious symptoms such as proteinuria.
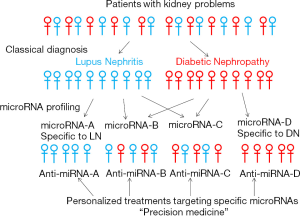
Accompanied by the development of recent technologies, the precise detection of miRNAs and suitable delivery methods are expected to be established. Hopefully, the diagnosis of patients by miRNA profiling and the treatment of patients by targeting specific miRNAs will be possible in the future for true personalized medicine (precision medicine) although much more efforts are necessary to identify miRNAs specific to diseases, stages or patients (individuals).
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Ming-Zhu Gao, MD (Department of Laboratory Medicine, Wuxi No.2 People’s Hospital of Nanjing Medical University, Wuxi, China).
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jlpm.2017.12.04). The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215-33. [Crossref] [PubMed]
- Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014;157:77-94. [Crossref] [PubMed]
- Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet 2015;16:421-33. [Crossref] [PubMed]
- Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell 2012;148:1172-87. [Crossref] [PubMed]
- de Planell-Saguer M, Rodicio MC. Analytical aspects of microRNA in diagnostics: a review. Anal Chim Acta 2011;699:134-52. [Crossref] [PubMed]
- Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008;18:997-1006. [Crossref] [PubMed]
- Etheridge A, Lee I, Hood L, et al. Extracellular microRNA: A new source of biomarkers. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis 2011;717:85-90. [Crossref] [PubMed]
- Trionfini P, Benigni A. MicroRNAs as Master Regulators of Glomerular Function in Health and Disease. J Am Soc Nephrol 2017;28:1686-96. [Crossref] [PubMed]
- Kato M, Natarajan R. Diabetic nephropathy--emerging epigenetic mechanisms. Nat Rev Nephrol 2014;10:517-30. [Crossref] [PubMed]
- Barutta F, Tricarico M, Corbelli A, et al. Urinary Exosomal MicroRNAs in Incipient Diabetic Nephropathy. PLoS One 2013;8:e73798 [Crossref] [PubMed]
- Szeto CC, Ching-Ha KB, Ka-Bik L, et al. Micro-RNA expression in the urinary sediment of patients with chronic kidney diseases. Dis Markers 2012;33:137-44. [Crossref] [PubMed]
- Wang G, Kwan BC, Lai FM, et al. Urinary sediment miRNA levels in adult nephrotic syndrome. Clin Chim Acta 2013;418:5-11. [Crossref] [PubMed]
- Yang Y, Xiao L, Li J, et al. Urine miRNAs: potential biomarkers for monitoring progression of early stages of diabetic nephropathy. Med Hypotheses 2013;81:274-8. [Crossref] [PubMed]
- Cai X, Xia Z, Zhang C, et al. Serum microRNAs levels in primary focal segmental glomerulosclerosis. Pediatr Nephrol 2013;28:1797-801. [Crossref] [PubMed]
- Higuchi C, Nakatsuka A, Eguchi J, et al. Identification of Circulating miR-101, miR-375 and miR-802 as Biomarkers for Type 2 Diabetes. Metabolism 2015;64:489-97. [PubMed]
- Pezzolesi MG, Satake E, McDonnell KP, et al. Circulating TGF-beta1-Regulated miRNAs and the Risk of Rapid Progression to ESRD in Type 1 Diabetes. Diabetes 2015;64:3285-93. [Crossref] [PubMed]
- Cardenas-Gonzalez M, Srivastava A, Pavkovic M, et al. Identification, Confirmation, and Replication of Novel Urinary MicroRNA Biomarkers in Lupus Nephritis and Diabetic Nephropathy. Clin Chem 2017;63:1515-26. [Crossref] [PubMed]
- Jia Y, Guan M, Zheng Z, et al. miRNAs in Urine Extracellular Vesicles as Predictors of Early-Stage Diabetic Nephropathy. Journal of Diabetes Research 2016;2016:10.
- Kato M, Natarajan R. MicroRNAs in diabetic nephropathy: functions, biomarkers, and therapeutic targets. Ann N Y Acad Sci 2015;1353:72-88. [Crossref] [PubMed]
- Zhong X, Chung ACK, Chen HY, et al. miR-21 is a key therapeutic target for renal injury in a mouse model of type 2 diabetes. Diabetologia 2013;56:663-74. [Crossref] [PubMed]
- Chau BN, Xin C, Hartner J, et al. MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Sci Transl Med 2012;4:121ra18 [Crossref] [PubMed]
- Long J, Wang Y, Wang W, et al. MicroRNA-29c is a signature microRNA under high glucose conditions that targets Sprouty homolog 1, and its in vivo knockdown prevents progression of diabetic nephropathy. J Biol Chem 2011;286:11837-48. [Crossref] [PubMed]
- Kato M, Putta S, Wang M, et al. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol 2009;11:881-9. [Crossref] [PubMed]
- Putta S, Lanting L, Sun G, et al. Inhibiting microRNA-192 ameliorates renal fibrosis in diabetic nephropathy. J Am Soc Nephrol 2012;23:458-69. [Crossref] [PubMed]
- Kato M, Wang M, Chen Z, et al. An endoplasmic reticulum stress-regulated lncRNA hosting a microRNA megacluster induces early features of diabetic nephropathy. Nat Commun 2016;7:12864. [Crossref] [PubMed]
- Badal SS, Wang Y, Long J, et al. miR-93 regulates Msk2-mediated chromatin remodelling in diabetic nephropathy. 2016;7:12076.
- van Rooij E, Kauppinen S. Development of microRNA therapeutics is coming of age. EMBO Mol Med 2014;6:851-64. [Crossref] [PubMed]
- Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017;16:203-22. [Crossref] [PubMed]
Cite this article as: Kato M. More microRNAs as biomarkers and hope for precision medicine in kidney diseases. J Lab Precis Med 2017;2:94.


