
Epigenetic implication in atrial fibrillation: a potential biomarker?
Introduction
Atrial Fibrillation (AF) is one of the gemstones in cardiovascular disease (CVD). It is the most studied arrhythmia in recent years. Everything about AF, from bench to bedside seems very interesting to be discussed (1). It leads to an evolving understanding of the pathogenesis and management of a patient with AF. However, further studies are required because AF is a very complex disease. The initiation and the perpetuation of AF caused by atrial electrical, cellular, and structural remodeling had been widely described (2). In contrary, we also know that AF has several risk factors. The understanding of how the risk factors causing cellular and electrical remodeling lead to the possible role of genetic influences in the mechanism of AF’s initiation (3). Although not popular as a genetic disorder, a small portion (5%) of all patients with AF is believed to be inherited. Few of familial cases of AF had been reported. Several genes and genetic loci had been identified to be responsible for the pathogenesis of AF (4). Another growing knowledge showed that environmental factors manipulated the gene expression and had a strong correlation with the genesis of AF, such as air pollution (5). Hopefully, by knowing and understanding gene and the system over it, we can identify and intervene early.
In the clinical perspective, AF is a devastating disease that has so many clinical manifestation and potential complications. Preventing and diagnosing the disease earlier is our general concern. Just like other disease, markers, or biomarkers play a potential role. Many biomarkers had been found to have a correlation with AF such as markers of inflammation that believed to have a major role in the genesis of AF (i.e., C-reactive protein), cardiac troponin and others. Although its position in recent guidelines not well established, the searching for perfect biomarkers continues (6).
The combining of the epigenetic process and its role as biomarkers is not very popular in the field of arrhythmia. However, in oncology (e.g., breast cancer) biomarkers that indicating DNA methylation starts to help the physician to prevent the disease, primary and secondary (7). This review will talk about the possible role of epigenetic biomarkers in AF.
What is epigenetic?
Epigenetic consists of two words, “epi” and “genetic”. Epigenetic is something above the genetic or on top of genetic (8). It was not a quite new and rare topic in the field of genetics. First, describe in the 1940s by Waddington, and today we may see thousands of pieces of literature that talk about this topic, spreading in many fields not only genetic and medicine but also ecology and other topics that may not be predicted before (9). The precise definition of epigenetic is difficult to be defined. Epigenetic is the process that controlled how the gene expressed without causing any changes in the DNA itself. The gene expression altered by turning the transcription process “on and off” (3,8). Epigenetic explain how the gene act, maintain, and interpret the information (10). There are several known mechanisms on how epigenetic work to control the gene, such as adding methyl components to the DNA structure, microRNA (miRNA) and modified the histone.
As a supra system of our genetic information, epigenetic had a major role in our body not only causing the disease but also to maintain normal condition (11). Epigenetic proven to be responsible in several disorders such as malignancy and other neurodegenerative processes. The epigenetic process caused alteration in gene function and causing cell malignancy. Malignancy is the first disease linked to the epigenetic mechanism. Cancer is believed to be a genetic disease for a long time, but its correlation with epigenetic first described when a study by Feinberg in 1983 showed hypomethylation of the DNA (12).
Epigenetic process
DNA methylation
DNA methylation can change the transcriptional process by adding a methyl group (-CH3) to the DNA (13). For example, adding methyl to cytosine (C4H5N3O) will change it to 5-methylcytosine. Which in turn will cause inactive gene expression, since only cytosine (not methylated one) that will be transcript to guanine. The methylation process will make methyl binding protein (MBP) prevent the transcriptional factor (TF) bind and proceed to the next transcriptional process.
Histones modification
Histones the are alkaline protein that pack the DNA so it can fit in the cell nucleus. Besides its function packing the DNA, it also has several post-translational modifications and regulates the gene expression. Histone can be methylated, acetylated, and other modification processes that can alter the gene expression (14). Histones H3 and H4 are the most obvious part of the histone that had a clear role in transcription regulation (15). For example, hyperacetylation by histone acetyltransferases (HATs) can activate the transcription process of TF (TAFII250). While deacetylation of histone will cause the gene silencing. Both of that process had a role in gene regulation (13,16).
miRNA
miRNA is an important gene expression regulator. miRNA is a small non-coding RNA. Its controlled gene expression by binding to messenger RNA (mRNA) and make match base pair. By binding with mRNA, it suppresses the mRNA translational process. Besides, it also can promote mRNA degradation through make a mismatch base pair. miRNA is a gene silencer. It normally presents in mammalian and believed to be the part of our evolution process by silencing some primordial gene (17). Besides, it presents in normal and physiological condition, miRNA found to be responsible for several disorders. In cancer, several miRNAs had been associated with the disease such as miR125b, miR145, miR21, miR155, miR210 in breast cancer; miR155, let7a in lung cancer; and miR29b in hepatocarcinoma. While miR144, miR146a, miR150, miR182, miR103, miR107 were found to be correlated with diabetes mellitus and miR30b, miR30c, miR26a, miR133b, miR184, let7 in neurodegenerative disease such as Parkinson (18).
Epigenetic in AF
It has been our common knowledge that heart disease development correlated with several risk factors, such as genetic factor, environmental factor (such as air pollution, exposure to chemical factors and fine particulate) and behavioral factors (19). However, how those factors collaborate and causing biomolecular alteration, initiating, and later, progress to heart disease is still unclear (20). The epigenetic process seems to be the best explanation of how those factors interact, although there is still a grey area that needs to be understood in the future.
DNA methylation and AF
Correlation between DNA methylation and other CVDs such as hypertension and atherosclerosis had been studied. In hypertension, the addition of methyl component to a specific DNA region that initiates the transcription process of 11 beta-hydroxysteroid dehydrogenases leads to blood pressure increasing (21). It showed how epigenetic role in hypertension. Other epigenetic process involved in the initiation of atherosclerosis process, by controlling multiple gene expressions such as cellular antioxidant superoxide dismutase (SOD), estrogen receptor (α and β), matrix metalloproteinase (MMP)-2, MMP-7, MMP-9, p53 protein and endothelial nitric oxide synthase (eNOS) (22). As we all know, those CVDs are known as the risk factor of AF. However, what about the direct epigenetic role to AF. Although the evidence not very clear, but the missing link between epigenetic and AF maybe through the cardiac fibrosis (3). Atrial wall fibrosis will lead to electrical, structural, and molecular remodeling that may be the initiator of the first AF episode (23).
In the experiment using rat, DNA methyltransferase 3A (DNMT3A) found to be significantly overexpressed in fibrotic myocardium. DNMT3A is one of the DNMT enzyme found in mammalian, the enzyme that catalyzes the methylation process. Increasing methylation process to Ras-association domain family 1 isoform A (RASSF1A) will cause repression of its function. As RASSF1A has a negative effect on ERK1/2 pathway, it is silencing due to the methylation process will increase cardiac fibrosis through that pathway (24). Other genes expression that altered by methylation process and prone to the development of AF are sulfonylurea receptor 2 (SUR2) overexpression (than SUR1), Pitx2 and sarco-endoplasmic reticulum calcium ATPase 2a (SERCA2a) (3,25).
Dietary intake (behavioral factors) which have rich methyl donors can lead to DNA changes (through DNA methylation) and prone to the development of CVD (20,26). The most popular proof of how dietary intake can increase the risk of CVD is in the Netherlands cohort epidemiology study. During Germany invasion on the 1940s, hunger is spreading all of the countries. Exposure to hunger causing the methylation in the developing fetal DNA. Moreover, in the future, baby that born in that period significantly had a higher number of metabolic and CVDs (27).
Histone modification and AF
The direct link between histone modification and AF can be clearly explained through the fibrotic process. Histone modification is linked with several kinds of fibrosis processes, such as in the liver, mesangial cell, and lung (28). On the other side, AF is believed to be initiated by some degree of atrial fibrosis (29).
As previously mentioned, histone deacetylase (HDAC) involved in histone modification (deacetylation). A study by Kee et al. in 2006 proved that HDAC inhibitors could inhibit cardiac myocyte hypertrophy that induced by administration of angiotensin II (30). Angiotensin II also had been linked to atrial fibrosis in another study (31). Although direct study to know the effect in AF is still ongoing, but it showed that histone modification inhibition might have a beneficial effect on AF.
miRNA and AF
Besides DNA methylation, miRNA is the epigenetic mechanism that becomes a point of AF’s researcher interest in the last decade. Many miRNAs had been detected to have a role in the development of the disease. miRNA1, miRNA26, miRNA133, miRNA328, miRNA499, and miRNA106b-25 had been correlated with atrial electrical remodeling and led to AF. They are known to have several alterations in AF, overexpressed, or suppressed (3).
For example, miRNA1 detected to be significantly lower in inpatient with chronic AF compared to sinus rhythm. miRNA1 and miRNA133 regulate gene function in pacemaker cell activity such as in hyperpolarization-activated cyclic nucleotide-gated (HCN) (32). It will enhance the activity of funny channel (If) in atrial myocyte and causing atrial ectopy that in turn will become the initiator of AF (33).
Suppression of miRNA106b will increase gene expression of ryanodine receptor (type 2). Ryanodine receptor had a major function in Ca2+ ion transporter within the cells. Increasing Ca2+ leak from sarcoplasmic reticulum has already known as the initiator of AF (34).
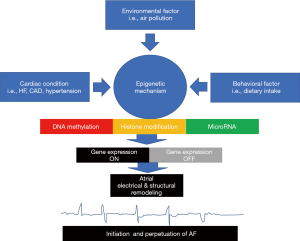
Another mechanism that involved in the genesis of AF is structural remodeling of the atrial wall. Several miRNA changes were identified, i.e., miRNA21, miRNA133, miRNA590, miRNA30, miRNA146b-5p, and miRNA206. Expression of miRNA133 correlated with the expression of TGF 1. Gene suppression of miRNA133 will increase the signaling pathway of TGF 1. TGF 1 signaling pathway had a major role in the AF’s structural remodeling by inducing atrial fibrosis (35). Figure 1 schematically summarizes the relation of some risk factors in epigenetic process of AF.
Epigenetic as a potential biomarker
What is epigenetic biomarker?
Any changes in the epigenetic mechanism (DNA methylation, histone modification, and mRNA) that can be detected in the tissue and body fluid is called epigenetic biomarker (36). Just like any other biomarkers, it should be had a role in diagnosing, predicting, and forecasting the disease outcome and the response to the therapy. There are some benefits of using epigenetic changes as a biomarker compared to other usual biomarkers. Epigenetic biomarkers can supply more individual gene functions that may be characterized by an environmental factor, diet, and habit. The study showed that epigenetic biomarker is stable in body fluid or tissue preparation. However, the standardized procedure had to be defined in further clinical trials, since Glinge in 2017 found that there is a decremental level of miRNA (miRNA21 and miRNA29b) after stored in room temperature (37).
Another thing that has to be considered is the cost since for screening cost-effectiveness is everything. In the future, epigenetic biomarkers seem to have a major role in theragnosis therapy. A biomarker that not only used as diagnostic tools but also had an important role in determining targeted therapy.
Available epigenetic biomarkers in the market
Recently, there were only two epigenetic biomarkers approved by FDA, i.e., Epi proColon and Cologuard. Both are used in colorectal carcinoma. It works by detecting DNA methylation (an epigenetic process) in septin 9, taken from a peripheral blood sample. This method showed high sensitivity and specificity 75–81% and 91–96%, respectively. However, it still not had a good position in the current guideline’s management of colorectal carcinoma. The European guideline did not mention any recommendation about using this method in inpatient screening. While some of the Chinese guidelines recommended using septin 9 in screening colorectal carcinoma (36).
Epigenetic biomarker in other CVD
Based on recent major guidelines, both European and American, there is no single recommendation to use epigenetic biomarkers in term of diagnostic and screening. Bringing it from bench to bedside, there is still a need for further studies and more robust evidence.
Coronary artery disease (CAD)
CAD is known to be related to the inflammation process. Interleukin-6 (IL-6) plays an important role in this process. Hypomethylation of IL-6 promoters has shown to increase the risk of atherosclerosis process (38). This methylation is caused by several previously known CAD risk factors, such as air pollution and methyl rich donor diet (20). Besides IL-6, another hypomethylation that related to atherosclerosis is long interspersed nuclear element-1 (LINE-1) hypomethylation. In the Chinese population, hypomethylation of LINE-1 associated with increased risk of CAD. With odds ratio ranging from 1.9–2.3 (39). Thus, it can be concluded that in the future, these two hypomethylation processes can be used as future biomarkers of CAD.
Circulating miRNA also can be used in the detection of CAD, since they both are correlated. Downregulation of miRNA155, miRNA145, miRNA17, miRNA126 and miRNA92a and up-regulation of miRNA133a, miRNA208a, and miRNA370 correlated with CAD through the inflammatory pathway.
However, none of those epigenetic biomarkers had been mentioned in the guideline’s recommendation.
Heart failure (HF)
Epigenomic mapping of the cardiomyopathic and normal heart showed a specific pattern. Platelet/endothelial cell adhesion molecule-1 (PECAM-1) promoter hypermethylation was observed in HF patients. Several pieces of DNA hypomethylation was also observed and known to reduce some gene expression in the failing heart. Such as methylation alteration in the lymphocyte antigen 75 (Ly75) promoter found in diabetic cardiomyopathy patient. Ly75 can affect cardiac myocyte contractility (40). Study in cancer had concluded that the epigenetic process had a major role in controlling the cell’s mitochondrial function. The mitochondrial function itself had a major role in the pathogenicity of HF (41). Several cellular metabolism intermediates such as NAD, β-hydroxybutyrate, ketoglutarate are found in HF patients. Those metabolic intermediate are the substrate for epigenetic changes and alter the signaling process and end up with pathological remodeling in the failing heart (40). If the DNA methylation and other epigenetic process are suspected to be the initiator of pathological changes in HF, its role in biomarkers is clear in predicting and screening. Besides that, it supposed to have a theragnostic role in this field.
Hypertrophic cardiomyopathy (HCM)
DNA methylation suppresses several genes that correlated with HCM, such as TNNI3 and BNP. Histone modification also detected in HCM, such as covalent modification (H3K9me3). Its role of targeted therapy was clear, but no evidence shows its role as diagnostic (biomarkers) tools (42).
How about epigenetic biomarkers in AF?
Biomarkers’ role in AF is receiving a new position in the guidelines. ESC guidelines 2016 mentioned that biomarkers could be used to determine the risk of stroke and bleeding risk in AF patients with the class recommendation of IIb and level of evidence of B (43). Previously, no recommendation is found despite many studies talking about it. High sensitivity troponin (Hs trop), natriuretic peptide, angiotensin II, and others are some of the biomarkers related to AF diagnostic (6). As mentioned previously, many potential biomarkers had been identified in the setting of AF. No epigenetic biomarkers had been validated to be used in the clinical setting until now. Table 1 lists some epigenetics alterations based on its mechanism.
Table 1
| DNA methylation | Histone modification | miRNA |
|---|---|---|
| Overexpression of DNMT3A | H3, H4 acetylation on MHC | miRNA1, 26, 133, 328, 499, 106b-25 (electrical remodeling) |
| PitX2c promoter methylation | Phosphorylation of HDAC5 | miRNA21, 133, 590, 30, 146b-5p, 206 (structural remodeling) |
| Methylation dependent on suppression of SUR2 | H3K9 demethylation | – |
| DNA methylation in the SERCA2a | – | – |
AF, atrial fibrillation; DNMT3A, DNA methyltransferase 3A; SUR2, sulfonylurea receptor 2; SERCA2a, sarco-endoplasmic reticulum calcium ATPase 2a; MHC, major histocompatibility complex; HDAC, histone deacetylase; miRNA, microRNA.
Barrier and future direction of epigenetic biomarker in AF
Although it seems to be very promising, the widely available clinical use of epigenetic biomarker still need a long way to go, and further studies need to be conducted if we want to include it in major guidelines recommendation of AF. Some major obstacles in this field also need to be addressed. Some gaps in the global understanding of epigenetic mechanisms had to be defined. Standardized procedures need to be established to prevent variability in the result. Evidence had been shown that different preparation would be had a different result. For example, in miRNA examination, there are extremely strict criteria that had to be met in order to give the perfect and standard result. Bringing laboratory results to the hospital for clinical use had some problems in the lab preparation. Technology such as new generation sequencing (NGS) and mass spectrometry need not only the high cost but also specific staff education (36). To be a biomarker, it should fit what is called “perfect biomarker”. The perfect biomarker should be detected as early as possible before the pathological process happened. It should be had high sensitivity and high specificity.
Moreover, for clinical use, it also had to be easily accessible, such as specimen that taken from peripheral blood flow is more clinically available than specimen from myocardial biopsy (44). What are some criteria that had to be met by the future epigenetic biomarkers? Another thing that had to be considered in order to become “biomarkers of choice”, epigenetic biomarker had to compete with existing and established biomarkers in AF such as cardiac troponin, atrial natriuretic peptide, and others. It had to give better performance. Figure 2 summarizes barriers on epigenetic applications.

Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Fabian Sanchis-Gomar) for the series “The Effects of Demographic Factors on Cardiovascular Biomarkers” published in Journal of Laboratory and Precision Medicine. The article has undergone external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jlpm.2019.09.02). The series “The Effects of Demographic Factors on Cardiovascular Biomarkers” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Fitzmaurice DA, Hobbs FR. Research into practice: management of atrial fibrillation in general practice. Br J Gen Pract 2014;64:540-2. [Crossref] [PubMed]
- Wijffels MC, Kirchhof CJ, Dorland R, et al. Electrical remodeling due to atrial fibrillation in chronically instrumented conscious goats. Circulation 1997;96:3710-20. [Crossref] [PubMed]
- Tao H, Shi KH, Yang JJ, et al. Epigenetic mechanisms in atrial fibrillation: New insights and future directions. Trends Cardiovasc Med 2016;26:306-18. [Crossref] [PubMed]
- Lubitz SA, Ozcan C, Magnani JW, et al. Genetics of atrial fibrillation. Circ Arrhythm Electrophysiol 2010;3:291-9. [Crossref] [PubMed]
- Link MS, Luttmann-Gibson H, Schwartz J, et al. Acute exposure to air pollution triggers atrial fibrillation. J Am Coll Cardiol 2013;62:816-25. [Crossref] [PubMed]
- Ardhianto P, Yuniadi Y. Biomarkers of atrial fibrillation: which one is a true marker? Cardiol Res Pract 2019;2019:8302326.
- Terry MB, McDonald JA, Wu HC, et al. Epigenetic biomarkers of breast cancer risk: across the breast cancer prevention continuum. Adv Exp Med Biol 2016;882:33-68. [Crossref] [PubMed]
- Rettner R. Epigenetics: definition & examples. Live Science. Available online: https://www.livescience.com/37703-epigenetics.html
- Deans C, Maggert KA. What do you mean, “epigenetic”? Genetics 2015;199:887-96. [Crossref] [PubMed]
- Olzscha H, Sheikh S, La Thangue NB. Deacetylation of chromatin and gene expression regulation: a new target for epigenetic therapy. Crit Rev Oncog 2015;20:1-17. [Crossref] [PubMed]
- Rasool M, Malik A, Naseer MI, et al. The role of epigenetics in personalized medicine: challenges and opportunities. BMC Med Genomics 2015;8:S5. [Crossref] [PubMed]
- Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 1983;301:89-92. [Crossref] [PubMed]
- Mazzio EA, Soliman KF. Basic concepts of epigenetics. Epigenetics 2012;7:119-30. [Crossref] [PubMed]
- de Cubas AA, Rathmell WK. Epigenetic modifiers: activities in renal cell carcinoma. Nat Rev Urol 2018;15:599-614. [Crossref] [PubMed]
- Freeman L, Kurumizaka H, Wolffe AP. Functional domains for assembly of histones H3 and H4 into the chromatin of Xenopus embryos. Proc Natl Acad Sci U S A 1996;93:12780-5. [Crossref] [PubMed]
- Solow S, Salunek M, Ryan R, et al. TAFII 250 phosphorylates human transcription factor IIA on serine residues important for TBP binding and transcription activity. J Biol Chem 2001;276:15886-92. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [Crossref] [PubMed]
- Li Y, Kowdley KV. MicroRNAs in common human diseases. Genomics Proteomics Bioinformatics 2012;10:246-53. [Crossref] [PubMed]
- Cosselman KE, Navas-Acien A, Kaufman JD. Environmental factors in cardiovascular disease. Nat Rev Cardiol 2015;12:627-42. [Crossref] [PubMed]
- Zhong J, Agha G, Baccarelli AA. The role of DNA methylation in cardiovascular risk and disease. Circ Res 2016;118:119-31. [Crossref] [PubMed]
- Friso S, Pizzolo F, Choi SW, et al. Epigenetic control of 11 beta-hydroxysteroid dehydrogenase 2 gene promoter is related to human hypertension. Atherosclerosis 2008;199:323-7. [Crossref] [PubMed]
- Turunen MP, Aavik E, Ylä-Herttuala S. Epigenetics and atherosclerosis. Biochem Biophys Acta 2009;1790:886-91.
- Everett TH, Olgin JE. Atrial fibrosis and the mechanisms of atrial fibrillation. Heart Rhythm 2007;4:S24-7. [Crossref] [PubMed]
- Tao H, Yang JJ, Chen ZW, et al. DNMT3A silencing RASSF1A promotes cardiac fibrosis through upregulation of ERK1/2. Toxicology 2014;323:42-50. [Crossref] [PubMed]
- Fatima N, Claycomb WC, Flagg TP. Promoter DNA methylation regulates murine SUR1 (Abcc8) and SUR2 (Abcc9) expression in HL-1 cardiomyocytes. PLoS One 2012;7:e41533 [Crossref] [PubMed]
- Ryan DP, Henzel KS, Pearson BL, et al. A paternal methyl donor-rich diet altered cognitive and neural functions in offspring mice. Mol Psychiatry 2018;23:1345-55. [Crossref] [PubMed]
- Lumey LH, Stein AD, Kahn HS, et al. Cohort profile: the Dutch hunger winter families study. Int J Epidemiol 2007;36:1196-204. [Crossref] [PubMed]
- Yao H-W, Li J. Epigenetic modifications in fibrotic diseases: implications for pathogenesis and pharmacological targets. J Pharmacol Exp Ther 2015;352:2-13. [Crossref] [PubMed]
- Platonov PG. Atrial fibrosis: an obligatory component of arrhythmia mechanisms in atrial fibrillation? J Geriatr Cardiol 2017;14:233-7. [PubMed]
- Kee HJ, Sohn IS, Nam KI, et al. Inhibition of histone deacetylation blocks cardiac hypertrophy induced by angiotensin II infusion and aortic banding. Circulation 2006;113:51-9. [Crossref] [PubMed]
- Novo G, Guttilla D, Fazio G, et al. The role of the renin–angiotensin system in atrial fibrillation and the therapeutic effects of ACE-Is and ARBS. Br J Clin Pharmacol 2008;66:345-51. [Crossref] [PubMed]
- Li YD, Hong YF, Yusufu AJ, et al. Altered expression of hyperpolarization-activated cyclic nucleotide-gated channels and microRNA-1 and -133 in patients with age-associated atrial fibrillation. Mol Med Rep 2015;12:3243-8. [Crossref] [PubMed]
- Stillitano F, Lonardo G, Giunti G, et al. Chronic atrial fibrillation alters the functional properties of If in the human atrium. J Cardiovasc Electrophysiol 2013;24:1391-400. [Crossref] [PubMed]
- Li N, Chiang DY, Wang S, et al. Ryanodine receptor-mediated calcium leak drives progressive development of an atrial fibrillation substrate in a transgenic mouse model. Circulation 2014;129:1276-85. [Crossref] [PubMed]
- Gramley F, Lorenzen J, Koellensperger E, et al. Atrial fibrosis and atrial fibrillation: the role of the TGF-β1 signaling pathway. Int J Cardiol 2010;143:405-13. [Crossref] [PubMed]
- García-Giménez JL, Seco-Cervera M, Tollefsbol TO, et al. Epigenetic biomarkers: Current strategies and future challenges for their use in the clinical laboratory. Crit Rev Clin Lab Sci 2017;54:529-50. [Crossref] [PubMed]
- Glinge C, Clauss S, Boddum K, et al. Stability of Circulating Blood-Based MicroRNAs - Pre-Analytic Methodological Considerations. PLoS One 2017;12:e0167969 [Crossref] [PubMed]
- Zuo HP, Guo YY, Che L, et al. Hypomethylation of interleukin-6 promoter is associated with the risk of coronary heart disease. Arq Bras Cardiol 2016;107:131-6. [PubMed]
- Wei L, Liu S, Su Z, et al. LINE-1 hypomethylation is associated with the risk of coronary heart disease in chinese population. Arq Bras Cardiol 2014;102:481-8. [PubMed]
- Kim SY, Morales C, Gillette TG, et al. Epigenetic regulation in heart failure. Curr Opin Cardiol 2016;31:255-65. [Crossref] [PubMed]
- Brown DA, Perry JB, Allen ME, et al. Mitochondrial function as a therapeutic target in heart failure. Nat Rev Cardiol 2017;14:238-50. [Crossref] [PubMed]
- Zhang QJ, Chen HZ, Wang L, et al. The histone trimethyllysine demethylase JMJD2A promotes cardiac hypertrophy in response to hypertrophic stimuli in mice. J Clin Invest 2011;121:2447-56. [Crossref] [PubMed]
- Kirchhof P, Benussi S, Kotecha D, et al. 2016 ESC Guidelines for the management of atrial fibrillation developed in collaboration with EACTS. Eur Heart J 2016;37:2893-962. [Crossref] [PubMed]
- Institute of Medicine. Qualifying Biomarkers. In: Institute of Medicine. Emerging safety science: workshop summary. Washington, DC: National Academies Press, 2008.
Cite this article as: Rizal A, Yuniadi Y. Epigenetic implication in atrial fibrillation: a potential biomarker? J Lab Precis Med 2019;4:33.


