
Innovative technologies for diagnosis and screening of genetic diseases in antenatal age
Introduction
Congenital anomalies occur in 2–4% of all newborns and cause 20.4% of perinatal deaths (1). Several technics have been developed and used over time to detect chromosomal alterations or single gene variants to optimize the reproductive outcome. Thanks to the advances in molecular biology methods, the number and quality of tests available for antenatal identification of genetic diseases greatly increased; the options now available range from non-invasive prenatal screening (NIPT) to targeted assessment of at-risk couples (2). In addition, the genetic carrier screening, an option prior to conception, allows each partners to obtain information about their genotype in relation to autosomal recessive, dominant or X-linked diseases; therefore, the purpose of genetic carrier screening is to plan following reproductive choices (i.e., traditional prenatal diagnosis (PND), preimplantation genetic diagnosis, egg or sperm donation or adoption).
Antenatal genetic tests aim to provide information on the genotype of the embryo [pre-implantation genetic testing (PGT)] or fetus (traditional PND, NIPT). These tests are voluntary and should only be undertaken after genetic counselling to inform about the nature of the test, the possible results and the options available (2). Antenatal diagnostic strategies are triggered by parents’ risk factors and/or genetic diseases previously identified in the family and/or ultrasound alterations recognized in the fetus. The outcome of preimplantation or prenatal genetic diagnosis is an informed reproductive decision-making starting from the analysis of family history, ethnic origin, past obstetric history and parents’ carrier status, up to molecular analysis on an embryonic or fetal biological sample (3). The most common genetic conditions for which antenatal genetic tests are required include cystic fibrosis, thalassemia, Duchenne/Becker muscular dystrophy and Fragile X syndrome: these tests allow to identify the genetic alterations previously described in the father and in the mother, therefore, an absence of those specific genetic alterations does not exclude the possibility that the child may have others (3).
It should be noted that prenatal diagnostic strategies have different aims than those of NIPT and genetic carrier screening. NIPT is applied to pregnant women, through the sequencing of maternal cell free DNA (cfDNA) and analysis of free fetal DNA (cffDNA), for the detection of fetal trisomy 13, 18, 21, sex chromosomes alterations, CNVs, and microdeletions (4).
This review will focus on analysing the tests in use when the reproductive path is already underway, with the aim of providing advice to optimize the molecular diagnosis for chromosomal and genetic conditions detectable in antenatal age thanks to the most innovative available technologies. The specialist in laboratory medicine and in health reproduction can benefit from this update, especially, when they have to manage couples who are planning a pregnancy, or who are pregnant, and couple at risk of genetic alteration transmission to the offspring with associated morbidity and/or mortality.
Methods
This review reflects emerging clinical and scientific advances as of the issued date and is subject to change. The paper takes into account the current scientific literature. In particular, Medline was conducted on PubMed, in a period between 2010 to 2019, using the following keywords: antenatal diagnosis, preimplantation diagnosis, PND, NIPT, antenatal age.
Antenatal genetic diagnosis
Antenatal genetic diagnosis is defined as a process that excludes or detects embryonic and fetal diseases. In addition, results provide information to parents and doctors focused on improving pregnancy outcomes and the well-being of the family (3) (Figure 1).
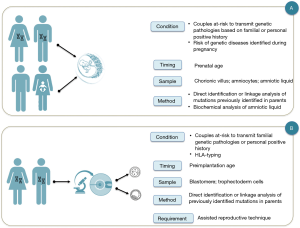
Since the termination of pregnancy was an option in case of a negative outcome of the prenatal genetic diagnosis, many efforts have been made to obtain a diagnosis as soon as possible: indeed, pre-implantation genetic diagnosis protocols have been developed as an alternative to traditional prenatal genetic diagnosis (3). In addition, an increasing number of prenatal and perinatal treatments are now available thanks to the development of in-utero therapies and the identification of pathologies sensitive to therapy during prenatal age or immediately after birth (5,6).
Diagnostic indications
Two main diagnostic indications from antenatal tests can be identified:
(I) patients or couples before the conception awareness about the risk of transmitting a genetic pathology to their offspring, since in their familial or personal medical history there are known factors that could cause genetically detectable pathologies, or even preventable. For example, couples with previous children affected by genetically determined disorders, couples in which one of the partners has blood relatives affected by genetically determined disorders, families of specific ethnic origin associated with an increased risk of abnormal pregnancy outcomes. In these cases, the couples can decide between the traditional PND (Figure 1A) and the pre-implantation genetic diagnosis (Figure 1B);
(II) patients or couples without known or previously recognizable risk factors, in which a routine blood test or a prenatal ultrasound scan reveals an unexpected abnormal finding during a routine pregnancy check (Figure 1A). Examples include cystic fibrosis mutation panel screening and CFTR gene sequencing for fetal echogenic bowel, FGFR gene sequencing for possible fetal achondroplasia or craniosynostosis, and targeted testing for mutations in genes causing genetic syndrome (7-11).
Family history and genetic counselling
The collection of family history is an essential tool for assessing the risk of genetic diseases, since the identification of risk factors related to genetic diseases and genetic diseases known in the family, allows the stratification of reproductive risk and the organization of targeted diagnostic approaches (2). Therefore, in all couples intending to have children, the family history should be carefully collected in order to identify the possible presence of hereditary genetic diseases in blood relatives. When the suspicion of a specific hereditary disease emerges from the family history, it would be necessary to deep investigate with genetic counselling.
Genetic counselling should be carried out, preferably in the preconception period, to partner/couples identified as being at risk of transmission of an inherited condition based on a three-generation pedigree review, ethnic background, personal manifestations or obstetrical/past medical history (intellectual disability, muscular dystrophy or bleeding disorders). Here, information on carrier screening and prenatal/preimplantation genetic diagnosis must be provided. Information on ethnic origin may be helpful in choosing appropriate screening studies. Direct gene mutation or expanded next-generation gene sequencing (NGS) testing should be discussed as part of the informed consent process.
Sampling techniques
A brief overview of the well-known conventional techniques is provided below, taking into account that, after fertilization, the zygote differs first in the blastomere and then in blastocyst, which contains an internal cellular mass that develops in the fetus and an external trophoblastic layer that develops in non-fetal structures such as the amnion, the chorion and the placenta (12,13). The genetic complement of the external cell mass almost always reflects the genetic constitution of the internal cell mass (i.e., the fetus) because both are derived from the same zygote.
Blastomere (6–8 cells) biopsy
Blastomere biopsy was the first technique developed for embryo sampling. It is carried out 3 days post-fertilization by aspirating 1–2 cells after the mechanical or chemical dissociation of the zona pellucida; therefore, in this case the sample for diagnosis is represented by a single cell or, occasionally, two (12).
Blastocyst (trophectoderm) biopsy
This sampling strategy is similar to blastomere biopsy but allows to obtain, 5 days post-fertilization, a sample with a greater number of cells (up to 10 cells) (12).
Chorionic villus sampling
Chorionic villus sampling allows to analyze fetal DNA between 11–14 weeks’ gestation. This procedure is carried out under ultrasound guidance, both transabdominally if the placenta is located anteriorly or transcervically if the placenta is positioned posteriorly, and is based on the sampling of chorionic villi. The DNA obtained from this fetal tissue, after maternal contamination exclusion, can be used for cytogenetics and molecular biology tests. Since it is an invasive procedure, a risk of miscarriage is present, being 1/400 (13).
Amniocentesis
Amniocentesis is carried out between 16–20 weeks’ gestation, but can be performed also later. This procedure is carried out under ultrasound guidance to collect amniotic fluid and then isolate fetal cells. Thus, it can be used for cytogenetics and molecular biology tests (analyzing the DNA of the isolated and cultured fetal cells), and also for biochemical evaluation directly on the amniotic fluid. In this case, the risk of miscarriage ranges from 1/500 to 1/900 (13).
Single gene disorders detectable by molecular analysis
Molecular analysis allows to identify an ever-increasing number of genetic pathologies in the antenatal age. The diagnostic strategy is based on the direct identification of the mutation or on the linkage analysis, in any case the prerequisite is the genotypic characterization of the parents and the family index case. Therefore, this type of investigation is indicated for couples in which there is a risk for a known genetic disorder. The molecular diagnosis must be preceded by a thorough genetic consultation, aimed at informing the couple about the implications of potential results (i.e., the result is relative to the specific pathology under examination, but does not exclude the presence of other genetic pathologies) as well as complications (spontaneous abortion related to sampling procedures, in the case of traditional PND) and requirements (use of assisted procreation techniques, even in couples able to conceive naturally) (3,8).
For couples at risk of transmitting a hereditary disorder specific to their offspring there are two diagnostic options, PGT or PND. These two diagnostic procedures share the same diagnostic purpose, but differ in diagnostic time, type of sampling and laboratory procedures (Figure 1). PGT has the advantage of anticipating the timing of the diagnosis thus avoiding the use of therapeutic abortion, but it presents the disadvantage to recognize as a complementary test not a substitute for PND (3).
Molecular techniques
Currently, the most recent PND strategies to detect chromosomal alterations, when the pregnancy is in progress, are based on echography coupled with karyotype and microarray (14). Karyotype, carried out by analyzing cultured fetal cells obtained from both chorionic villus sampling or amniocentesis, allows to analyze all the 23 pairs of chromosomes, including sex chromosomes. Therefore, the advantage is that it allows to identify chromosomal alterations, such as trisomy, monosomy, large deletions and duplications, translocations, inversions, and mosaic; while, the disadvantages are that it requires cell culture, the time of the analysis is long (up to 2 weeks).
Microarray-based chromosomal evaluation has shown a higher sensitivity; thus, it is recommended as a first-level test when ultrasound alterations are detected (15). This technic allows the identification of chromosomal aneuploidy, microdeletions and microduplications within a chromosome not detectable by a standard karyotype due to their size. Several platforms can be used for chromosome microarray including the array-based comparative genomic hybridization (aCGH). Even if the balanced chromosomal translocations are not detectable, the microarray platforms present some advantages such as the absence of cells culture and reduction of the turnaround time of the diagnostic test (16). The introduction of microarray-based procedures has increased the diagnostic yield of prenatal tests respect to traditional cytogenetics (17,18). However, 70–80% of fetus with ultrasound alterations still remain without a molecular diagnosis (17,18).
The above-mentioned cytogenetic strategies cannot detect alterations at single nucleotide level. As a consequence, in the case of the inherited disease (based on familial history and/or ultrasound specific features, as shown in Figure 1), molecular biology techniques can be used to analyze specific disease-causing genes and highlight the causative mutations already identified in the family (8,9). To this aim direct sequencing of the exon(s) harboring the familial mutation(s) or STRs analysis are used in routine settings (8,9,19). However, also in this case the test may be not informative (8,9,19). These limitations are expected in both prenatal and preimplantation genetic diagnosis (3).
The limits of, currently used, traditional prenatal diagnostic strategies have fueled the need for more sensitive techniques; the recent development of novel sequencing technologies is prompting PND into a new era (20). Indeed, NGS, as for other fields of molecular diagnostics, is changing PND: in addition to targeted sequencing of single disease-causing genes or small group of genes of interest, prenatal exome sequencing (ES) is now being an option, at least in selected cases; genome sequencing has been used for research purposes but is not yet applicable in clinical settings, due to difficulties into data interpretation; finally, RNA-sequencing may provide new insights into human development during pregnancy, and novel non-invasive biomarkers to monitor placental functions through maternal plasma cell-free RNA (cfRNA) (2,20-23). Considering all the above, it is expected that NGS-based approach, to direct analyze fetal specimens from amniocentesis and chorionic villus sampling, will replace soon more traditional prenatal methodologies.
The most simple application of NGS is the analysis of single disease-causing genes starting from fetal DNA. This approach can be advantageous when the disease present within a family is well defined based on clinical/biochemical features, but the mutation(s) are not known in the parents.
An effective approach in the case of multigenic inheritance or confounding, overlapping phenotypes, is the analysis of multi-genes panels. Targeted multi-genes panels allow to simultaneously analyze a group of genes related to a disease or to a group of diseases sharing common phenotypic features, or a common genetic background. This approach is now widely used for the diagnosis of several diseases in postnatal settings and has been also applied to PND (24,25). However, even if targeted multi-genes panel analysis allows to optimize cost and time of the genetic tests, it may be ineffective if the choice of the panel to be tested is guided by the phenotype, and the latter (based just on ultrasound) is not clearly defined. Another possible use of multi-genes panels in the prenatal medicine is the so called “carrier screening”. This test, differently from the others, does not analyze fetal DNA but is performed on the parents (preferably before conception) to verify if they carry recessive alterations that may be risky if transmitted to their newborn (13). Indeed, since carriers of autosomal recessive disorders are generally healthy and often have also a negative family history, they may be not aware regarding their status as “carriers”.
ES is currently used in clinical setting to provide postnatal diagnosis and has showed its utility in increasing the diagnostic rate of complex phenotype (26,27). Its use in PND is still at embryonal stage (28-30). The advantage of ES is that, by analyzing all the coding genes in the human genome without selection biases, it may identify a molecular alteration also in the cases where other targeted approaches fail. However, the huge amount of data produced is still challenging to interpret in routine application (31). Nevertheless, several reports are showing the utility of prenatal ES in selected case by using trio analysis in order to facilitate variants filtering, prioritization and interpretation, and are also expanding our knowledge on prenatal phenotype of such rare inherited diseases (28,29,32). Currently, the cost, difficulties into variants interpretation, missing of phenotypic information, and the ethical considerations, limits ES use in routine being rather a second level test for selected, difficult cases (20).
Whole genome sequencing (WGS) has been used in prenatal settings as proof of principle, however its feasibility is lower than ES since not only costs are higher, but the difficulties related to variants interpretation are exponentially increased due to the inclusion also of the non-coding genomic regions (33).
Finally, RNA-Seq studies on fetal tissues are still rare (20). This approach has the potential to provide novel insights into fetal development and clarify the mechanisms underlying specific diseases of interest, however currently may be useful just for research purposes and is limited by scarce tissues availability.
NIPT
The NIPT, performed in the last 30 years, are essentially based on the analysis of biochemical markers on maternal blood, combined with ultrasound investigations (Figure 2).
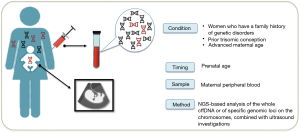
The free fetal DNA (cffDNA) analysis present in the maternal blood was analyzed for the first time in the 1997 when the presence of the Y chromosome in the plasma of some women with male fetuses was revealed. The analysis of cffDNA is the innovative prenatal screening for chromosomal abnormalities, CNVs and microdeletion.
Diagnostic indications
Since the techniques actually used for NIPT are based on the analysis of the total circulating DNA, without differentiating the fetal from the maternal one, NIPT is not be considered a diagnostic test but a screening test.
Nowadays the NIPT is well validated for chromosomal anomalies (such as trisomies of chromosomes 21, 18, 13), sex chromosome aneuploidies, CNVs and microdeletions, however the detection of single-gene disorders using cffDNA is the real challenge (3).
Despite accredited scientific societies suggest the NIPT to all pregnant women, it is recommended to women who have a family history of genetic disorders, women who have previously had a baby with a chromosomal abnormality, or who are over 35—the cut-off for “advanced maternal age”.
Sampling techniques
NIPT is considered without risks for miscarriage, unlike the sampling of amniotic fluid or chorionic villi, as it is based on the analysis of cffDNA. It can be isolated early starting from week 10th of gestation, when it reaches sufficient amounts for potential clinical use. Its percentage can vary between <4%, a quantity not useful for the diagnosis, and about 40%; the percentage of cffDNA is called “fetal fraction” (34).
The cffDNA originates from the lysis of maternal and placental cells and for this reason it is degraded in fragments containing about 180 base pairs (bp) and are suspended in arterial plasma. The cffDNA present in maternal plasma originates from the placental cytotrophoblast and it is detectable yet at 5 weeks of pregnancy. Taking in to account the origin of cffDNA in a percentage of cases a possible to find a discordant karyotype with respect to the fetal one. The chromosomal analyzes of the trophoblast have, in fact, identified variable percentages of discordance for the different aneuploidies (34).
Molecular techniques
Non-invasive prenatal testing (NIPT) is the ensemble of molecular strategies that allow the screening, testing, or diagnosis of fetal chromosomal or genetic alterations by analyzing directly cffDNA in maternal plasma or serum. NGS has been successfully applied also to NIPT (35,36). In particular, NIPT coupled with NGS has been used for chromosomal aneuploidies detection using both whole fetal DNA sequencing (shotgun approach) or targeted sequencing of a panel of chromosomes more likely to give aneuploidies (chromosomes 13, 18, X and Y chromosomes).
In general, NIPT laboratory protocols are based on comparisons between a specific chromosome of analyzed sample with a disomic control that could be an internal control, another chromosome, or a pool of disomic pregnancies. The ratio between the counts (number of CR fragments in the test/number of fragments in the disomic reference samples) is approximately equal to 1.
If a fetus bearing trisomy 21 is present in the examined pregnancy, the FF increases due to the presence of additional circulating fragments released by the supernumerary CR21 of the fetus. The magnitude of the increase depends on the percentage of the total FF and on the number of bp of CR21, in relation to the bp of the overall genome of the fetus. Some NIPT tests insert the percentage of FF in the algorithm for the formulation of the probability of presence of the investigated trisomy, while others use predetermined normalization factors, which can still reach high levels of reliability (37).
As mentioned above, three main techniques for NIPT are based on NGS: NGS of the entire genome; NGS of specific regions; SNPs analysis, i.e., detection of polymorphisms of single nucleotides.
The NGS-based analysis of the whole cffDNA present in the maternal plasma generates millions of short sequences covering the entire genome, which are then mapped onto a reference sequence of the human genome, to establish their origin and count the number of fragments originating from the chromosome of interest, compared to the number of fragments obtained from the other chromosomes (38). Several algorithms have been developed to determine if, in the sample under examination, the number of fragments has increased or decreased, in relation to a threshold suggestive of an aneuploidy (39,40).
An alternative strategy selectively amplifies specific genomic loci on the chromosome(s) of interest, which are then sequenced. This technique is cheaper, as it reduces the regions to be sequenced, but has the limitation of studying only a few pre-selected regions of interest (37).
The third technique is based on the amplification of several loci polymorphic (SNP) on chromosome(s) of interest (41).
The sensitivity and specificity towards the main aneuploidies are high for all three techniques, regardless of the instruments used for sequencing and the bioinformatic algorithm used (42).
The most current researches are focused to develop a unique protocol able to analyze chromosomal abnormality, CNVs and monogenic diseases (43). In this context, NGS coupled with a specific bioinformatics pipeline for the analysis of the results become essential. The genetic disorders actually studied are Phenylketonuria and Wilson disease using circulating single molecule amplification and resequencing technology (cSMART) (44,45). The genotyping seems a reliable method for some recessive disease such as DMD (46). Recently, encouraging data showed the high analytical sensitivity and specificity by combining NGS with the quantitative counting of the template (input DNA) for the NIPT of the most common genetic disorders in the world (sickle cell disease, cystic fibrosis, spinal muscular atrophy, alpha-thalassemia, and beta-thalassemia) (47).
Finally, the recent possibility to study cell free amniotic RNA and other non -invasive approaches through the study of maternal cell free RNA will prompt this area of research (29).
Conclusions
The technical progresses and reduced sampling risks will make the techniques for antenatal diagnosis and screening increasingly effective in optimizing the reproductive outcome. The impact will be a reduced frequency of rare and serious hereditary genetic diseases and may, for example, lead to a negative selection of the most common late-onset genetic variants or recessive disorders (i.e., cystic fibrosis).
In the future, new diagnostic technologies will not only provide the tools to give prospective parents the opportunity to make an informed choice, but they can also lead to personalized fetal medicine. Antenatal genetic testing and genetic screening have many advantages, but also important technical challenges; moreover, the acceptance and integration of this diagnostic possibility paves the way for ethical-social debates.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Giuseppe Lippi, Martina Montagnana and Zhi-De Hu) for the series “Laboratory Medicine in Pregnancy” published in Journal of Laboratory and Precision Medicine. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jlpm.2019.11.02). The series “Laboratory Medicine in Pregnancy” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Murphy SL, Mathews TJ, Martin JA, et al. Annual summary of vital statistics: 2013-2014. Pediatrics 2017; [Crossref] [PubMed]
- Cariati F, D'Argenio V, Tomaiuolo R. The evolving role of genetic tests in reproductive medicine. J Transl Med 2019;17:267. [Crossref] [PubMed]
- Cariati F, Savarese M, D'Argenio V, et al. The SEeMORE strategy: single-tube electrophoresis analysis-based genotyping to detect monogenic diseases rapidly and effectively from conception until birth. Clin Chem Lab Med 2017;56:40-50. [Crossref] [PubMed]
- Vanstone M, Cernat A, Majid U, et al. Perspectives of Pregnant People and Clinicians on Noninvasive Prenatal Testing: A Systematic Review and Qualitative Meta-synthesis. Ont Health Technol Assess Ser 2019;19:1-38. [PubMed]
- Regan F, Lees CC, Jones B, et al. Prenatal Management of Pregnancies at Risk of Fetal Neonatal Alloimmune Thrombocytopenia (FNAIT): Scientific Impact Paper No. 61. BJOG 2019;126:e173-85. [Crossref] [PubMed]
- Simpson JL, Rechitsky S. Prenatal genetic testing and treatment for congenital adrenal hyperplasia. Fertil Steril 2019;111:21-3. [Crossref] [PubMed]
- Tomaiuolo R, Fausto M, Elce A, et al. Enhanced frequency of CFTR gene variants in couples who are candidates for assisted reproductive technology treatment. Clin Chem Lab Med 2011;49:1289-93. [Crossref] [PubMed]
- Tomaiuolo R, Nardiello P, Martinelli P, et al. Prenatal diagnosis of cystic fibrosis: an experience of 181 cases. Clin Chem Lab Med 2013;51:2227-32. [Crossref] [PubMed]
- Fuccio A, Iorio M, Amato F, et al. A novel DHPLC-based procedure for the analysis of COL1A1 and COL1A2 mutations in osteogenesis imperfecta. J Mol Diagn 2011;13:648-56. [Crossref] [PubMed]
- Saal HM. Genetic Evaluation for Craniofacial Conditions. Facial Plast Surg Clin North Am 2016;24:405-25. [Crossref] [PubMed]
- González-Huerta NC, Valdés-Miranda JM, Pérez-Cabrera A, et al. Noonan syndrome: prenatal diagnosis in a woman carrying a PTPN11 gene mutation. J Matern Fetal Neonatal Med 2010;23:688-91. [Crossref] [PubMed]
- Schmutzler AG. Theory and practice of preimplantation genetic screening (PGS). Eur J Med Genet 2019;62:103670 [Crossref] [PubMed]
- Krstić N, Običan SG. Current landscape of prenatal genetic screening and testing. Birth Defects Res 2019; [Epub ahead of print]. [Crossref] [PubMed]
- Wellesley D, Dolk H, Boyd PA, et al. Rare chromosome abnormalities, prevalence and prenatal diagnosis rates from population-based congenital anomaly registers in Europe. Eur J Hum Genet 2012;20:521-6. [Crossref] [PubMed]
- Committee on Practice Bulletins—Obstetrics, Committee on Genetics, and the Society for Maternal-Fetal Medicine. Practice bulletin No. 163: Screening for Fetal aneuploidy. Obstet Gynecol 2016;127:e123-37. [Crossref] [PubMed]
- Gardner RM, Sutherland GR, Shaffer LG. Chromo-some abnormalities and genetic counseling. 4th ed. New York, USA: Oxford University Press, 2011.
- Hillman SC, McMullan DJ, Hall G, et al. Use of prenatal chromosomal microarray: prospective cohort study and systematic review and meta-analysis. Ultrasound Obstet Gynecol 2013;41:610-20. [Crossref] [PubMed]
- Shaffer LG, Rosenfeld JA, Dabell MP, et al. Detection rates of clinically significant genomic alterations by microarray analysis for specific anomalies detected by ultrasound. Prenat Diagn 2012;32:986-95. [Crossref] [PubMed]
- Elce A, Boccia A, Cardillo G, et al. Three novel CFTR polymorphic repeats improve segregation analysis for cystic fibrosis. Clin Chem 2009;55:1372-9. [Crossref] [PubMed]
- Vora NL, Hui L. Next-generation sequencing and prenatal 'omics: advanced diagnostics and new insights into human development. Genet Med 2018;20:791-9. [Crossref] [PubMed]
- Nunziato M, Esposito MV, Starnone F, et al. A multi-gene panel beyond BRCA1/BRCA2 to identify new breast cancer-predisposing mutations by a picodroplet PCR followed by a next-generation sequencing strategy: a pilot study. Anal Chim Acta 2019;1046:154-162. [Crossref] [PubMed]
- Bergougnoux A, D'Argenio V, Sollfrank S, et al. Multicenter validation study for the certification of a CFTR gene scanning method using next generation sequencing technology. Clin Chem Lab Med 2018;56:1046-53. [Crossref] [PubMed]
- D'Argenio V, Esposito MV, Telese A, et al. The molecular analysis of BRCA1 and BRCA2: Next-generation sequencing supersedes conventional approaches. Clin Chim Acta 2015;446:221-5. [Crossref] [PubMed]
- Hu P, Qiao F, Wang Y, et al. Clinical application of targeted next-generation sequencing on fetuses with congenital heart defects. Ultrasound Obstet Gynecol. 2018;52:205-11. [Crossref] [PubMed]
- Rasmussen M, Sunde L, Nielsen ML, et al. Targeted gene sequencing and whole-exome sequencing in autopsied fetuses with prenatally diagnosed kidney anomalies. Clin Genet 2018;93:860-9. [Crossref] [PubMed]
- Fang YL, Zhang RP, Wang YZ, et al. A novel mutation in a common pathogenic gene (SETD5) associated with intellectual disability: A case report. Exp Ther Med 2019;18:3737-40. [PubMed]
- Liaqat K, Hussain S, Bilal M, et al. Further evidence of involvement of TMEM132E in autosomal recessive nonsyndromic hearing impairment. J Hum Genet 2019; [Epub ahead of print]. [PubMed]
- Jelin AC, Vora N. Whole exome sequencing: applications in prenatal genetics. Obstet Gynecol Clin North Am 2018;45:69-81. [Crossref] [PubMed]
- Vora NL, Powell B, Brandt A, et al. Prenatal exome sequencing in anomalous fetuses: new opportunities and challenges. Genet Med 2017;19:1207-16. [Crossref] [PubMed]
- Best S, Wou K, Vora N, et al. Promises, pitfalls and practicalities of prenatal whole exome sequencing. Prenatal Diagnosis 2018;38:10-9. [Crossref] [PubMed]
- D'Argenio V. The High-Throughput Analyses Era: Are We Ready for the Data Struggle? High Throughput 2018. doi:
10.3390/ht7010008 . - Fu F, Li R, Li Y, et al. Whole exome sequencing as a diagnostic adjunct to clinical testing in fetuses with structural abnormalities. Ultrasound Obstet Gynecol 2018;51:493-502. [Crossref] [PubMed]
- Mao Q, Chin R, Xie W, et al. Advanced whole-genome sequencing and analysis of fetal genomes from amniotic fluid. Clin Chem 2018;64:715-25. [Crossref] [PubMed]
- Ashoor G, Syngelaki A, Poon LC, et al. Fetal fraction in maternal plasma cell-free DNA at 11-13 weeks' gestation: relation to maternal and fetal characteristics. Ultrasound Obstet Gynecol 2013;41:26-32. [Crossref] [PubMed]
- Wong FC, Lo YM. Prenatal Diagnosis Innovation: Genome Sequencing of Maternal Plasma. Annu Rev Med 2016;67:419-32. [Crossref] [PubMed]
- Lo YM. Non-invasive prenatal testing using massively parallel sequencing of maternal plasma DNA: from molecular karyotyping to fetal whole-genome sequencing. Reprod Biomed Online 2013;27:593-8. [Crossref] [PubMed]
- Zhang H, Gao Y, Jiang F, et al. Non-invasive prenatal testing for trisomies 21, 18 and 13: clinical experience from 146,958 pregnancies. Ultrasound Obstet Gynecol 2015;45:530-8. [Crossref] [PubMed]
- Ong FS, Lin JC, Das K, et al. Translational utility of next-generation sequencing. Genomics 2013;102:137-9. [Crossref] [PubMed]
- Hudecova I, Sahota D, Heung MM. Maternal plasma fetal DNA fractions in pregnancies with low and high risks for fetal chromosomal aneuploidies. PLoS One 2014;9:e88484 [Crossref] [PubMed]
- Tiller GE, Kershberg HB, Goff J, et al. Women's views and the impact of noninvasive prenatal testing on procedures in a managed care setting. Prenat Diagn 2015;35:428-33. [Crossref] [PubMed]
- Stokowski R, Wang E, White K, et al. Clinical performance of non-invasive prenatal testing (NIPT) using targeted cell-free DNA analysis in maternal plasma with microarrays or next generation sequencing (NGS) is consistent across multiple controlled clinical studies. Prenat Diagn 2015;35:1243-6. [Crossref] [PubMed]
- van Beek DM, Straver R, Weiss MM, et al. Comparing methods for fetal fraction determination and quality control of NIPT samples. Prenat Diagn 2017;37:769-73. [Crossref] [PubMed]
- Luo Y, Jia B, Yan K, et al. Pilot study of a novel multi-functional noninvasive prenatal test on fetus aneuploidy, copy number variation, and single-gene disorder screening. Mol Genet Genomic Med 2019;7:e00597 [Crossref] [PubMed]
- Duan H, Liu N, Zhao Z, et al. Non-invasive prenatal testing of pregnancies at risk for phenylketonuria. Arch Dis Child Fetal Neonatal Ed 2019;104:F24-9. [Crossref] [PubMed]
- Lv W, Wei X, Guo R, et al. Noninvasive prenatal testing for Wilson disease by use of circulating single-molecule amplification and resequencing technology (cSMART). Clin Chem 2015;61:172-81. [Crossref] [PubMed]
- Xu Y, Li X, Ge HJ, et al. Haplotype-based approach for noninvasive prenatal tests of Duchenne muscular dystrophy using cell-free fetal DNA in maternal plasma. Genet Med 2015;17:889-96. [Crossref] [PubMed]
- Tsao DS, Silas S, Landry BP, et al. A novel high-throughput molecular counting method with single base-pair resolution enables accurate single-gene NIPT. Sci Rep 2019;9:14382. [Crossref] [PubMed]
Cite this article as: Cariati F, D’Argenio V, Tomaiuolo R. Innovative technologies for diagnosis and screening of genetic diseases in antenatal age. J Lab Precis Med 2020;5:6.


