Assessment of immune response against SARS-CoV-2 at emergency department evaluation in COVID-19 patients
As the coronavirus disease 2019 (COVID-19) pandemic soldiers on, serology testing has yet to find a clearly defined role in the diagnostic and prognostic paradigm for clinical management of this condition (1). The rapid development of a vast number of immunoassays with limited rigorous and independent validation studies has led to a wide range of reported sensitivities and specificities, limiting the utility of such tests from a diagnostic perspective (1). Furthermore, our ability to interpret the results of these assays has been plagued by a poor understanding of the immune response to this novel pathogen. A recent Dutch clinical trial evaluating convalescent plasma for reducing COVID-19 mortality was abruptly stopped due to high titers of neutralizing antibodies in the majority of sick patients at the time of study inclusion despite their clinical status (2). Recent evidence also suggests a substantial decline in antibody levels after severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, further complicating our understanding of the immune response against this virus (3,4).
Many investigations have used time from symptom onset to evaluate antibody response, but this approach suffers from recall bias. Therefore, we report here on anti-SARS-CoV-2 specific antibodies at presentation to the Emergency Department (ED), in addition to self-reported time from symptom onset. As a clinical time point, ED presentation reflects the moment when a patient felt the need to seek immediate medical care, either due to initial or progressive decline in clinical status. Therefore, serology status at that time point may have important therapeutic or prognostic implications. We used two rigorously validated assays, one for detecting anti-SARS-CoV-2 IgA and the other one for anti-SARS-CoV-2 IgG. We also report total immunoglobulin levels (IgA, IgG, and IgM) at ED presentation and compare these levels between patients requiring and not-requiring intensive care unit (ICU) admission during the course of their illness.
Under an Institutional Review Board-approved waiver of informed consent, patients ≥18 years of age presenting to the University of Cincinnati Medical Center (UCMC) ED in April and May 2020 with at least one symptom consistent with COVID-19 (e.g., cough, fever, shortness of breath, loss of smell) and requiring bloodwork for clinical care were prospectively enrolled into the study. Importantly, this time period is when the initial wave of COVID-19 moved through the city. Only known prisoners were excluded. Blood samples were collected, centrifuged at 2,000 g for 15 minutes at 4 °C within 3 hours of collection, and frozen at −80 °C until analysis. Only patients with positive results of reverse transcription polymerase chain reaction (RT-PCR) nasopharyngeal swab obtained for clinical purposes were included in the study. Anti-SARS-CoV-2 IgA was measured using an enzyme-linked immunosorbent assay (ELISAs; Euroimmun AG, Luebeck, Germany). Anti-SARS-CoV-2 S1/S2 IgG was measured using chemiluminescence immunoassay (CLIA) on LIASON XL (DiaSorin S.p.A. Saluggia, Italy). Due to the reported variability and poor response to SARS-CoV-2 for IgM, we did not measure this antibody class (1). Total immunoglobulin levels (IgA, IgG, and IgM) were measured with a Behring Nephelometer II System (BN II, Siemens Medical Solutions USA, Inc., Malvern, PA, USA). Results of all assays were interpreted according to manufacturers’ recommendations. We compared levels between patients requiring and not-requiring ICU admission during the course of their illness. Categorical values were compared using Fischer’s exact test, while continuous variables were compared using Mann-Whitney U test, with P value <0.05 considered statistically significant for all analyses. This study was performed in compliance with the Declaration of Helsinki and local and national regulations.
A total of 52 patients met inclusion and exclusion criteria and were enrolled into the study. Due to aliquot limitations, only 35 patients could be tested for anti-SARS-CoV-2 IgA. The median age was 50.5 years [interquartile range (IQR): 39.3–66.0 years], and 42.3% (22/52) were females. The median self-reported time from symptom onset to ED presentation was 7 days (IQR: 3–10 days). While hospitalized, 28.8% (15/51) required ICU admission.
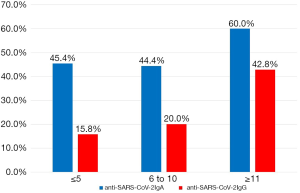
A summary of serology testing is shown in Table 1. At ED presentation, 45.7% and 25.0% of patients were positive for anti-SARS-CoV-2 IgA and anti-SARS-CoV-2 IgG, respectively. Seropositivity by days since self-reported symptom onset is presented in Figure 1. Interestingly, 45.4% of patients presenting at 5 days or less from symptom onset were positive for anti-SARS-CoV-2 IgA. No difference in the rate of seropositivity for either anti-SARS-CoV-2 antibody was observed between patients requiring ICU support vs. no ICU support. For total immunoglobulins, values above the upper limit of the reference range were seen in 5.8% (n=3) for IgM, 21.2% (n=11) for IgA, and 23.1% (n=12) for IgG, respectively. Although no statistically significant differences were observed between ICU and non-ICU patients for any of these immunoglobulins, a trend towards lower levels of IgM and IgG were observed among ICU patients.
Table 1
| Variable | All patients (n=52) | ICU (n=15) | No ICU (n=37) | P value |
|---|---|---|---|---|
| Euroimmun anti-SARS-CoV-2 IgA positive: n/total (%)* | 16/35 (45.7%) | 5/11 (45.4%) | 11/24 (45.8%) | 0.999 |
| DiaSorin anti-SARS-CoV-2 IgG positive: n/total (%) | 13 (25.0%) | 3 (20.0%) | 10 (27.0%) | 0.733 |
| Total IgA (mg/dL): median (IQR) | 285.5 (194.5–379.5) | 296.0 (196.0–370.0) | 277.0 (189.5–416.0) | 0.917 |
| Total IgM (mg/dL): median (IQR) | 92.4 (68.7–132.3) | 81.8 (49.1–109.0) | 96.3 (72.2–140.5) | 0.115 |
| Total IgG (mg/dL): median (IQR) | 1,250 (1,070–1,610) | 1,170 (1,070–1,250) | 1,390 (1,125–1,755) | 0.063 |
*, only 35 out of 52 patients were measured for IgA at admission. ED, emergency department; ICU, intensive care unit; IQR, interquartile range.
In summary, nearly half of the patients in our symptomatic cohort were positive for anti-SARS-CoV-2 IgA at time of ED evaluation, whilst a quarter were positive to anti-SARS-CoV-2 IgG, as the initial wave of COVID-19 moved through Cincinnati, USA. The rate of anti-SARS-CoV-2 positivity was not associated with need for ICU support. We also observed a rather high rate of anti-SARS-CoV-2 IgA positivity in patients early in their disease course. Lippi et al. (5), using the same anti-SARS-CoV-2 IgA ELISA kit, reported only 3.3% of patients with positive anti-SARS-CoV-2 IgA at ≤5 days as compared to 45.4% we observed, but this may reflect differences in study design and inclusion criteria (e.g., many of the patients included in the previous study were pre-symptomatic or asymptomatic). Notably, between 6–10 days, Lippi and colleagues (5) reported a seropositivity for IgA of 31%, which is more similar to that observed in our study (i.e., 44%). It is important to note that seropositivity does not necessarily indicate the presence of neutralizing antibodies or protective immunity, but only the presence of contact with SARS-CoV-2. Finally, in this cohort, total immunoglobulin levels and rate of anti-SARS-CoV-2 positivity at time of ED evaluation were not predictive of need for ICU support. However, a non-significant trend towards lower total IgM and IgG counts was observed in patients, which may be related to the lymphopenia commonly observed in patients with severe COVID-19 (6). This should be further addressed by future studies.
This study was limited by its relatively small sample size and single-center design. As noted earlier, any analysis with respect to self-reported symptom duration may suffer from potential recall bias. Finally, total immunoglobulin levels could be influenced by many variables, including patient comorbidities. Such confounders should be controlled for in larger studies assessing the adaptive immune response to COVID-19.
The results of our study further highlight the concept that serology testing should not be used as a surrogate for direct RNA identification in diagnosing acute SARS-CoV-2 infection, especially at ED evaluation. Relying on serology by assaying either IgA or IgG would generate an unacceptable number of false negative test results, which may contribute to the risk of propagating COVID-19 within healthcare facilities.
Acknowledgments
Funding: This study was funded by the University of Cincinnati College of Medicine Special Coronavirus (COVID-19) Research Pilot Grant Program.
Footnote
Provenance and Peer Review: This article was a free submission to the journal. The article did not undergo external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jlpm-20-87). GL serves as the unpaid Editor-in-Chief of the Journal of Laboratory and Precision Medicine from November 2016 - October 2021. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lippi G, Henry BM, Sanchis-Gomar F, et al. Updates on laboratory investigations in coronavirus disease 2019 (COVID-19). Acta Biomed 2020;91:e2020030 [PubMed]
- Gharbharan A, Jordans CCE, GeurtsvanKessel C, et al. Convalescent Plasma for COVID-19. A randomized clinical trial. medRxiv 2020. doi:
10.1101/2020.07.01.20139857 - Long QX, Tang XJ, Shi QL, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med 2020;26:1200-4. [Crossref] [PubMed]
- Seow J, Graham C, Merrick B, Acors S, et al. Longitudinal evaluation and decline of antibody responses in SARS-CoV-2 infection. medRxiv 2020. doi:
10.1101/2020.07.09.20148429 - Lippi G, Salvagno GL, Pegoraro M, et al. Assessment of immune response to SARS-CoV-2 with fully automated MAGLUMI 2019-nCoV IgG and IgM chemiluminescence immunoassays. Clin Chem Lab Med 2020;58:1156-9. [Crossref] [PubMed]
- Henry BM, de Oliveira MHS, Benoit S, et al. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin Chem Lab Med 2020;58:1021-8. [Crossref] [PubMed]
Cite this article as: Benoit JL, Benoit SW, Rose J, Hoehn J, Lippi G, Henry BM. Assessment of immune response against SARS-CoV-2 at emergency department evaluation in COVID-19 patients. J Lab Precis Med 2020;5:33.