
The real origin of SARS-CoV-2: does it really matter?
The ongoing coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is still spreading across the world, infecting several millions of people and causing over a million deaths to-date (1). COVID-19 is the third coronavirus outbreak that has affected humanity during the past two decades, preceded by severe acute respiratory syndrome (SARS) in 2002–2003 and Middle-East respiratory syndrome (MERS) in 2012 (2). As with all other coronaviruses, SARS-CoV-2 is an enveloped virus with positive-sense, single-stranded RNA genome, encoding four leading structural proteins which are known as Spike [S; containing the receptor-binding domain (RBD) through which the virus binds to its natural receptors at the surface of host cells], the envelope protein (E), the membrane protein (M) and the nucleocapsid protein (N) (3).
There is an ongoing discussion, which often fuels fierce debates, about the real origin of SARS-CoV-2 (4). We personally proffer that scientific debate as to whether this “new” coronavirus has been created, or reengineered, within a research or clinical laboratory in Wuhan or elsewhere all around the world is virtually meaningless. Nonetheless, the mysterious SARS-CoV-2 origin is now arguably used by many scientists and policymakers as a form of political rhetoric, for the poorly hidden purpose of destabilizing economic and geopolitical relationships (5). Establishing whether SARS-CoV-2 is humanmade or not seems virtually impossible, unless someone were to openly admit that the virus has been entirely fabricated, modified or has leaked from a laboratory.
Leaving political speculations aside from real science, several lines of evidence garnered since the beginning of this pandemic now allow to credibly pinpoint that SARS-CoV-2 has a “zoonotic” animal origin, a conclusion supported by a deep analysis of the genome and evolutionary history of this virus. A bat is indeed the original source of SARS-CoV-2, as well as of many other life-threatening viruses such as Ebola, rabies, Influenza A and so forth. A phylogenetic analysis with structural modeling of spike protein has revealed that this moiety has a 97% sequence identity with that of another bat coronavirus (i.e., BatCov-RaTG13), and also shares a 97% sequence identity in the RBD with a pangolin coronavirus [i.e., 2019 Guandong (GD) Pangolin] (6). This may suggest that SARS-CoV-2 has probably originated from recombination of these two animal coronaviruses in an intermediate host (e.g., pangolin?), and has then infected humans as a consequence of spillover. Notably, SARS-CoV-2 also shares 76% structural homology with SARS-CoV-1, the virus which caused the SARS outbreak nearly 20 years ago. However, the structural homology between these two viruses in the RBD is limited, lower than 50%, which would perhaps explain the different biological and clinical features between SARS and COVID-19. One of the most intriguing aspects in SARS-CoV-2 biology, is why this and the two previous similar coronaviruses that caused SARS and MERS display such high virulence and pathogenicity compared to other coronaviruses which only cause a common cold. A possible explanation has been given in the seminal work of Gussow et al. (7), in which it was demonstrated that these three coronaviruses have undergone a convergent trend in their evolution, characterized by enhancement of nuclear localization signal in their N protein, which contributes to modify their subcellular localization, combined with important insertions in the spike protein, which may ultimately facilitate and amplify their binding to host cell receptors. Another intriguing aspect in SARS-CoV-2 biology is that this virus undergoes considerable intra- and inter-human recombination. There is little doubt that SARS-CoV-2 has undergone many mutations since its first appearance in China and will continue to do so while it remains among us. This is not surprising since viruses which directly encode their genome in RNA, including HIV and influenza along with SARS-CoV-2, seems to insert mutations in their RNA rapidly. This occurs because these microorganisms reproduce inside their hosts, where enzymes copying RNA are more vulnerable to errors (8). A comprehensive description of SARS-CoV-2 phylogeny is available on the website of the Global Initiative on Sharing All Influenza Data (GISAID), which lists the huge complexity of the recombinant events that the virus has undergone thus far (a temporal resolution analysis is consistent with nucleotide replacement rate of around 8×104 yearly subs per site, giving rise to as many as 4,771 genomes sampled to date) (9). Interestingly, a study published by Tiwari and Mishra concluded that nonsynonymous substitutions generated not less than 57 amino acid changes distributed over different viral proteins, with maximum genetic variation observed in the region encoding for the spike protein, which may then be reflected by import structural and functional heterogeneity when the spike protein interacts with host cell receptors (10). The leading source of these mutations is hence human recombination. In the interesting study of Shen and colleagues, the median number of intra-host SARS-CoV-2 variants was found to range between 1 and 4 in infected patients, but cumulatively spanning over a very vast array, between 0 and 51 (11). Rather understandably, this intra-individual recombination process may have considerable impacts on virulence, infectivity, pathogenicity and transmissibility of the virus.
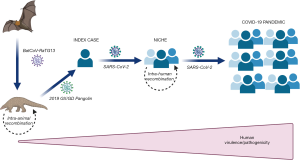
Conspiracy theories on the origin and spread of COVID-19 have ranged from the virus being a lab-designed bioweapon or 5G cell phone towers which accelerate the spread of the virus or a meteor which carried the virus to planet Earth when it struck northeastern China in October (12). Such theories have also been rapidly spread around the world via social media. Nonetheless, the current biological evidence would lead us to generate a possible theory on the origin and spillover of SARS-CoV-2, as shown in Figure 1. Briefly, the virus has likely originated from a bat coronavirus, probably BatCoV-RaTG13, which has been transmitted to another intermediate animal, maybe a pangolin, where the ancestral virus has undergone primary intra-animal recombination. This new virus (e.g., 2019 GX/GD Pangolin) was likely then transmitted to the first human index case, where it then became the SARS-CoV-2 that originated in the first local outbreak. Failure to timely recognize the first diffusion of the virus within a likely circumscribed human niche has enabled further intra-human recombination processes, which then finally generated the highly virulent pathogen that is causing the ongoing pandemic outbreak. This theory is inherently supported by data showing that the virus has circulated at lower virulence much earlier before its first identification in Wuhan. For example, evidence from environmental monitoring suggests that the virus started to circulate in Italy as early as December 2019, while the much larger outbreak was only recorded 2–3 months later (13).
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was a standard submission to the journal. The article did not undergo external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jlpm-20-94). GL serves as the unpaid Editor-in-Chief of the Journal of Laboratory and Precision Medicine from November 2016–October 2021. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lippi G, Sanchis-Gomar F, Henry BM. Coronavirus disease 2019 (COVID-19): the portrait of a perfect storm. Ann Transl Med 2020;8:497. [Crossref] [PubMed]
- Lippi G, Sanchis-Gomar F, Henry BM. COVID-19: unravelling the clinical progression of nature’s virtually perfect biological weapon. Ann Transl Med 2020;8:693. [Crossref] [PubMed]
- Lippi G, Mattiuzzi C, Bovo C, et al. Current laboratory diagnostics of coronavirus disease 2019 (COVID-19). Acta Biomed 2020;91:137-45. [PubMed]
- Lasco G. Medical populism and the COVID-19 pandemic. Glob Public Health 2020;15:1417-29. [Crossref] [PubMed]
- Tanne JH. Covid-19: Trump is criticised for again promoting unorthodox medical information. BMJ 2020;370:m3046. [Crossref] [PubMed]
- Jaimes JA, Andre NM, Chappie JS, et al. Phylogenetic Analysis and Structural Modeling of SARS-CoV-2 Spike Protein Reveals an Evolutionary Distinct and Proteolytically Sensitive Activation Loop. J Mol Biol 2020;432:3309-25. [Crossref] [PubMed]
- Gussow AB, Auslander N, Faure G, et al. Genomic determinants of pathogenicity in SARS-CoV-2 and other human coronaviruses. Proc Natl Acad Sci U S A 2020;117:15193-9. [Crossref] [PubMed]
- Grubaugh ND, Hanage WP, Rasmussen AL. Making Sense of Mutation: What D614G Means for the COVID-19 Pandemic Remains Unclear. Cell 2020;182:794-5. [Crossref] [PubMed]
- Global Initiative on Sharing All Influenza Data. Genomic epidemiology of hCoV-19. Available online: https://www.gisaid.org/epiflu-applications/hcov-19-genomic-epidemiology/. Accessed September 25 2020.
- Tiwari M, Mishra D. Investigating the genomic landscape of novel coronavirus (2019-nCoV) to identify non-synonymous mutations for use in diagnosis and drug design. J Clin Virol 2020;128:104441. [Crossref] [PubMed]
- Shen Z, Xiao Y, Kang L, et al. Genomic Diversity of Severe Acute Respiratory Syndrome-Coronavirus 2 in Patients With Coronavirus Disease 2019. Clin Infect Dis 2020;71:713-20. [Crossref] [PubMed]
- Steele EJ, Gorczynski RM, Lindley RA, et al. Origin of new emergent Coronavirus and Candida fungal diseases-Terrestrial or cosmic? Adv Genet 2020;106:75-100. [Crossref] [PubMed]
- La Rosa G, Mancini P, Bonanno Ferraro G, et al. SARS-CoV-2 has been circulating in northern Italy since December 2019: Evidence from environmental monitoring. Sci Total Environ 2021;750:141711. [Crossref] [PubMed]
Cite this article as: Lippi G, Henry BM, Sanchis-Gomar F. The real origin of SARS-CoV-2: does it really matter? J Lab Precis Med 2021;6:9.


