
The landscape of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genomic mutations
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is an enveloped virus with a positive-sense, single-stranded RNA genome, which extends over 29,903 bp (1). The virus, which was originally isolated in the Chinese city of Wuhan at the end of the 2019, has dominated the world, causing nearly 6 million deaths so far.
The common assumption that coronaviruses would be relatively less vulnerable to incorporate mutations in their genome, mostly supported by the presence of efficient proofreading pathways (2,3), is mostly unfounded and has now been refuted by facts. A paradigmatic example of an RNA virus that strongly modified its genome over time is the Influenza A virus H1N1, which has caused around 50 million deaths approximately one century ago (4), and has since incorporated more than 1,400 substitutions (i.e., over 10% of its entire genome), including more than 300 non-synonymous mutations (5). It is hence not surprising that SARS-CoV-2 has become subjected to a sustained selective pressure, resulting in emergence of many nucleotide substations and, accordingly, new variants, some of which are so fit that they are capable to progressively replace the former, trailing a path towards endemicity (6). SARS-CoV-2 accumulates mutations as its genome is copied inside the host cells. Reliable evidence has been published that a considerable number of intra-host variants can originate in each infected subject (i.e., up to 51) (7), that intra-host single nucleotide variations develop in the vast majority of infected people (i.e., over 80%) (8), and that the number of substitutions is directly dependent on the length of active infection (9). Therefore, the aim of this article is to provide an updated landscape of SARS-CoV-2 genomic mutations emerged since its first identification and sequencing, in 2019.
Methods
We downloaded an up-to-date list of all mutations detected within the SARS-CoV-2 RNA genome and submitted up to February 8, 2022 to the website of the National Center for Biotechnology Information (NCBI) (10). Briefly, this NCBI database contains all variants in Sequence Read Archive (SRA) records compared to the prototype SARS-CoV-2 reference sequence NC_045512.2 (SARS-CoV-2 isolate Wuhan-Hu-1, complete genome: 29903 bp ss-RNA) (1), with the purpose of detecting single nucleotide polymorphisms (SNPs) and related protein effects. The SRA runs are included when they reach at least 100 hits for SARS-CoV-2 through SARS-CoV-2 Detection Tool, with read length of ≥75. Insertion-deletion mutations (i.e., indels) were excluded. This study was conducted in accordance with Helsinki Declaration (as revised in 2013), under terms of relevant local legislation.
Statistical analysis
All mutations throughout the SARS-CoV-2 genome were downloaded from the NCBI database into a Microsoft Excel file (Microsoft Corp., Redmond, WA, USA), where they were both statistically and graphically analyzed.
Results
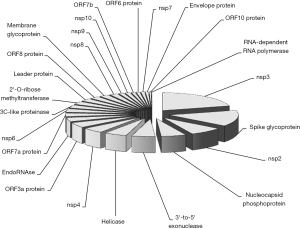
The search in the NCBI Mutations in SARS-CoV-2 SRA repository allowed to identify a total number of 26,005 different mutations up to February 8, 2022. The largest number of mutations, over 20%, could be located within the gene encoding for the Nsp3 protein (sequence 2,720–8,554), followed by the gene encoding for the Spike (S) protein with 14.6% (sequence 21,563–25,384). The number of mutations found in the other genes was considerably lower, as shown in Table 1, Figure 1 and website: https://cdn.amegroups.cn/static/public/jlpm-22-17-1.xls. Overall, 17,948/26,005 (69.0%) of these mutations interested single nucleotide positions, thus spanning over ~62% of the entire SARS-CoV-2 genome. Of all these mutations, 61.5% (15,999/26,005) were non-synonymous and the remaining synonymous. A specific analysis of mutations occurred within the gene encoding for the spike protein revealed that 2,299/3,799 (60.5%) were non-synonymous, whilst 661/3,799 (17.4%) involved the receptor binding domain (RBD; sequence 22,514–23,185), 59.2% of which (391/661) were non-synonymous. When the number of mutations was expressed as a ratio to the size of the relative gene, the highest ratio was found in the sequence encoding for the ORF7a protein (ratio, 2.25), followed by ORF7b (ratio, 1.85), ORF8 protein (ratio, 1.60) and ORF3a protein (ratio, 1.48). Notably, the gene encoding for the RNA-dependent RNA polymerase (RdRp) accounted for only 0.1% of all mutations reported in the NCBI database for the SARS-CoV-2 genome, with a considerably low ratio with gene size (i.e., ratio, 0.01) (Table 1).
Table 1
| SARS-CoV-2 protein | Genomic location | Gene size | Number of mutations | % of all mutations | Ratio mutations/gene size |
|---|---|---|---|---|---|
| 2'-O-ribose methyltransferase | 20,659–21,552 | 893 | 712 | 2.7 | 0.80 |
| 3C-like proteinase | 10,055–10,972 | 917 | 730 | 2.8 | 0.80 |
| 3'-to-5' exonuclease | 18,040–19,620 | 1,580 | 1,413 | 5.4 | 0.89 |
| EndoRNAse | 19,621–20,658 | 1,037 | 963 | 3.7 | 0.93 |
| Envelope protein | 26,245–26,472 | 227 | 185 | 0.7 | 0.81 |
| Helicase | 16,237–18,039 | 1,802 | 1,375 | 5.3 | 0.76 |
| Leader protein | 266–805 | 539 | 644 | 2.5 | 1.19 |
| Membrane glycoprotein | 26,523–27,191 | 668 | 568 | 2.2 | 0.85 |
| nsp2 | 806–2,719 | 1,913 | 2,006 | 7.7 | 1.05 |
| nsp3 | 2,720–8,554 | 5,834 | 5,393 | 20.7 | 0.92 |
| nsp4 | 8,555–10,054 | 1,499 | 1,276 | 4.9 | 0.85 |
| nsp6 | 10,973–11,842 | 869 | 819 | 3.1 | 0.94 |
| nsp7 | 11,843–12,091 | 248 | 223 | 0.9 | 0.90 |
| nsp8 | 12,092–12,685 | 593 | 491 | 1.9 | 0.83 |
| nsp9 | 12,686–13,024 | 338 | 322 | 1.2 | 0.95 |
| nsp10 | 13,025–13,441 | 416 | 314 | 1.2 | 0.75 |
| Nucleocapsid phosphoprotein | 28,274–29,533 | 1,259 | 1,512 | 5.8 | 1.20 |
| ORF10 protein | 29,558–29,674 | 116 | 130 | 0.5 | 1.12 |
| ORF3a protein | 25,393–26,220 | 827 | 1,225 | 4.7 | 1.48 |
| ORF6 protein | 27,202–27,387 | 185 | 230 | 0.9 | 1.24 |
| ORF7a protein | 27,394–27,759 | 365 | 823 | 3.2 | 2.25 |
| ORF7b protein | 27,756–27,887 | 131 | 242 | 0.9 | 1.85 |
| ORF8 protein | 27,894–28,259 | 365 | 584 | 2.2 | 1.60 |
| RNA-dependent RNA polymerase | 13,442–16,236 | 2,794 | 26 | 0.1 | 0.01 |
| Spike glycoprotein | 21,563–25,384 | 3,821 | 3,799 | 14.6 | 0.99 |
Discussion
The results of our analysis clearly demonstrate that SARS-CoV-2 has drastically mutated since its first sequence has been identified over 2 years ago (1), accumulating over 26,000 mutations, two-third of which were non-synonymous and thus associated with a change in the resulting protein that is expressed. The vast majority of all mutations (over one-third; 35.4%) could be identified in the sequences encoding for the Nsp3 protein and the spike glycoprotein. This is not really surprising, since both these two viral proteins play a substantial role in the virulence of SARS-CoV-2. Nsp3 is the largest protein encoded in the SARS-CoV-2 genome and is deeply involved in viral replication, since it forms a complex with Nsp 4 and Nsp 6, driving the modification of endoplasmic reticulum of the host cells into double membrane vesicles (DMVs) (11). DMVs are small-size vesicles (i.e., between 100–300 pm), typically delimited by two membranes, which generates an important scaffold for viral replication-transcription complexes (RTCs), enabling efficient protection against the host antiviral response (12). Another important function of Nsp3 entails its papain-like protease (PLpro) activity, which is effective to actively cleave Nsp1 from Nsp2, Nsp2 from Nsp3 and Nsp3 from Nsp4, thus ultimately promoting the release of the mature proteins (Nsp1, Nsp2 and Nsp3) from the viral polypeptide.
The spike protein is another essential component of SARS-CoV-2 virulence, in that its sequence would determine the affinity to its natural host cell receptor [i.e., angiotensin converting enzyme 2 (ACE2)], as well as with the major cofactors that modulate its infective potential [i.e., transmembrane protease serine 2 (TMPRSS2), furin, heparan sulphate proteoglycans, phosphatidylserine receptor, Neuropilin-1, CD147, C-type lectins] (13). It is hence reasonable that this important surface glycoprotein would be subjected to an extremely high evolutionary pressure, since sequence mutations in this domain may promote more stable receptor binding, better fusion with host cell plasma membrane and, ultimately, more efficient cell penetration, but may also be responsible for higher escape from the host immune system (14), as recently seen with the recently emerged Omicron (B.1.1.529) variant (15). Interestingly, 17.4% of spike protein mutations were located within the RBD, with a synonymous mutation rate that was marginally but non-statistically higher than that found in the rest of the SARS-CoV-2 genome (40.8% vs. 38.4%; Chi-square with Yates’ correction, P=0.110).
As concerns genes with higher mutation rate compared to their size, Orf7a displays a remarkably high value of 2.25 (Table 1), thus exhibiting a disproportionate number of mutations compared to its size (i.e., 823/365). Evidence suggests that Orf7a, a type I transmembrane protein, may have an active role in fostering budding and release of mature SARS-CoV-2 viral particles. Specifically, ORF7a seems to promote more efficient extracellular release of virions, by antagonizing the antiviral effect of type-I interferons and bone marrow stromal antigen 2 (BST-2/tetherin), which secure budding virions to host cells (11,13). The gene encoding for ORF7b, the second in terms of mutation rate compared to its size (242/131; ratio, 1.85), generates another transmembrane protein whose function remains elusive. Speculations have been made that this protein may interfere with some cellular pathways, ultimately promoting apoptosis of infected cells (11).
Conclusions
The results of our analysis of the NCBI database demonstrate that SARS-CoV-2 has incorporated a remarkable number of mutations since its emergence, such that many others are likely to emerge in the foreseeable future, at least while viral circulation remains higher and widespread. Proper monitoring will require major efforts to sequence the virus and timely identify important mutations that may affect it biological behavior and will turn into new lineages that may become variants of concern or have enhanced virulence and/or pathogenicity.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://jlpm.amegroups.com/article/view/10.21037/jlpm-22-17/coif). GL serves as the Editor-in-Chief of Journal of Laboratory and Precision Medicine from November 2016 to October 2023. The other author has no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature 2020;579:265-9. [Crossref] [PubMed]
- Manzanares-Meza LD, Medina-Contreras O. SARS-CoV-2 and influenza: a comparative overview and treatment implications. Bol Med Hosp Infant Mex 2020;77:262-73. [Crossref] [PubMed]
- Callaway E. The coronavirus is mutating - does it matter? Nature 2020;585:174-7. [Crossref] [PubMed]
- Sampath S, Khedr A, Qamar S, et al. Pandemics Throughout the History. Cureus 2021;13:e18136. [PubMed]
- Carter RW, Sanford JC. A new look at an old virus: patterns of mutation accumulation in the human H1N1 influenza virus since 1918. Theor Biol Med Model 2012;9:42. [Crossref] [PubMed]
- Lippi G, Mattiuzzi C, Henry BM. Updated picture of SARS-CoV-2 variants and mutations. Diagnosis (Berl) 2021;9:11-7. [Crossref] [PubMed]
- Shen Z, Xiao Y, Kang L, et al. Genomic Diversity of Severe Acute Respiratory Syndrome-Coronavirus 2 in Patients With Coronavirus Disease 2019. Clin Infect Dis 2020;71:713-20. [Crossref] [PubMed]
- Pathak AK, Mishra GP, Uppili B, et al. Spatio-temporal dynamics of intra-host variability in SARS-CoV-2 genomes. Nucleic Acids Res 2022;50:1551-61. [Crossref] [PubMed]
- Voloch CM, da Silva Francisco R Jr, de Almeida LGP, et al. Intra-host evolution during SARS-CoV-2 prolonged infection. Virus Evol 2021;7:veab078.
- National Center for Biotechnology Information. Mutations in SARS-CoV-2 SRA Data. Available online: https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/scov2_snp. Last accessed, February 8, 2022.
- Mariano G, Farthing RJ, Lale-Farjat SLM, et al. Structural Characterization of SARS-CoV-2: Where We Are, and Where We Need to Be. Front Mol Biosci 2020;7:605236. [Crossref] [PubMed]
- Wong LH, Edgar JR, Martello A, et al. Exploiting Connections for Viral Replication. Front Cell Dev Biol 2021;9:640456. [Crossref] [PubMed]
- Redondo N, Zaldívar-López S, Garrido JJ, et al. SARS-CoV-2 Accessory Proteins in Viral Pathogenesis: Knowns and Unknowns. Front Immunol 2021;12:708264. [Crossref] [PubMed]
- Evans JP, Liu SL. Role of host factors in SARS-CoV-2 entry. J Biol Chem 2021;297:100847. [Crossref] [PubMed]
- Lippi G, Mattiuzzi C, Henry BM. Neutralizing potency of COVID-19 vaccines against the SARS-CoV-2 Omicron (B.1.1.529) variant. J Med Virol 2022; Epub ahead of print. [Crossref] [PubMed]
Cite this article as: Lippi G, Henry BM. The landscape of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genomic mutations. J Lab Precis Med 2022;7:10.


