Using IPDmada to perform statistical analyses of diagnostic accuracy in primary studies: explanation and elaboration with a case study
Introduction
In a study of diagnostic test accuracy (DTA), one or more tests (referred to as the index tests) are evaluated for their ability in detecting or predicting a target condition or health status (1). The index test results are compared with a reference standard, which is the best available method for determining the target condition. Based on this comparison between the index test and the reference standard, diagnostic accuracy can be quantified with many performance measures, including but not limited to sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), positive likelihood ratio (LR+), negative likelihood ratio (LR−), diagnostic odds ratio (DOR), and areas under receiver operating characteristic (ROC) curve (AUC) (2,3).
The statistical analyses producing the above results can be done in many software, such as SPSS, Stata, SAS, and R. However, researchers need to calculate all those performance measures step-by-step. It is preferable to have an easy-to-use tool that requests minimum work by the users (e.g., only uploading the data) and provides all the results needed in a DTA study.
IPDmada (4) is web-based R Shiny application recently developed for individual patient data meta-analysis (IPD-MA) of DTA. IPDmada provides a wide range of analyses in IPD-MA, which are also applicable in primary studies of diagnostic accuracy. In this explanation and elaboration document, we use IPDmada to perform both basic and advanced statistical analyses in evaluating a diagnostic test to show the feasibility of applying IPDmada in primary studies of DTA.
Data
The data source used in this case study is from a previous publication by Norton et al., which investigated the diagnostic accuracy of transient evoked otoacoustic emissions (TEOAEs), distortion product otoacoustic emissions (DPOAEs), and auditory brain stem responses (ABRs) for neonatal hearing impairment (5).
The dataset is publicly available and can be downloaded from the website of Fred Hutchinson Cancer Research Center, Diagnostic and Biomarkers Statistical (DABS) Center (6), via this link: https://research.fredhutch.org/content/dam/stripe/diagnostic-biomarkers-statistical-center/files/nnhs2.csv.
The public dataset contains 5,058 records from 2,742 individuals in 6 centers, and the variables and their description are given in Table 1.
Table 1
| Variables | Description | Value |
|---|---|---|
| id | Patient ID | |
| ear | Ear | 1= left, 2= right |
| sitenum | Center ID | |
| currage | Corrected age | |
| gender | Gender | 1= female, 2= male |
| d | Hearing impaired | 0= no, 1= yes |
| y1 | DPOAE 65 at 2 kHz | |
| y2 | TEOAE 80 at 2 kHz | |
| y3 | ABR |
TEOAE, transient evoked otoacoustic emission; DPOAE, distortion product otoacoustic emission; ABR, auditory brain stem response.
Statistical analysis
Since IPDmada requests data in a specific format (i.e., CSV format), and with pre-defined variable names (i.e., Study, disease, test.results), data preparation is needed before uploading raw data to IPDmada.
In this illustration case study, data preparation is performed in spreadsheets, which can be done with any spreadsheet software, e.g., Microsoft Excel, Apple Numbers, Google Sheets, and WPS Spreadsheet, etc.
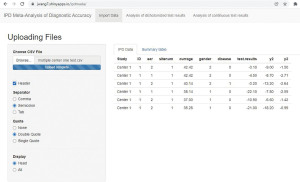
When data is ready for use, it will be uploaded to IPDmada (https://jwang7.shinyapps.io/ipdmada/), and all further analyses will be done automatically by IPDmada. Results can be downloaded either in tables (as CSV files) or figures (as PDF files).
The statistical analyses provided in IPDmada include:
- Test performance measures calculated from confusion matrix:
- True positive (TP), false negative (FN), false positive (FP), and true negative (TN);
- Sensitivity and specificity;
- PPV and NPV;
- LR+ and LR−;
- DOR.
Three options are provided to determine the positivity threshold, which is used to generate the confusion matrix: - Different thresholds are used across centers;
- Optimal thresholds are determined for each center (based on Youden’s Index);
- One specific threshold is used in all centers.
- Test performance based on the distribution of test results:
- Distribution of test results in disease and non-disease groups;
- ROC curve and AUC;
- Distribution of covariates in disease and non-disease groups;
- Covariate adjusted ROC curve and covariate-adjusted AUC.
Without loss of generality, in this case study we only show the analyses of TEOAEs, and the same analyses can be applied to DPOAEs and ABRs as well. Please note that, all the analyses presented in this report are only for illustration purposes, and sometimes variables are analyzed as a hypothetical example without practical meaning, thus no clinical conclusions should be drawn from these results.
Case study
Analyses of data from multiple centers for one test
In a multiple centers study, diagnostic data are collected from N centers. In this case, the data structure is similar with an IPD-MA including N primary studies. A single center study, or a study treating data from different centers as if from one single center, can be seen as a special case of a multiple centers study with N=1.
Data preparation
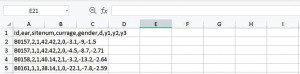
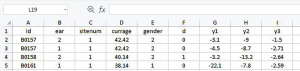
We start the data preparation from the CSV file downloaded from the website given in the Data section. Depending on the software, data can be read in as comma-separated values (Figure 1) or columns (Figure 2). Data in the form of Figure 1 can be transformed into Figure 2 with the Convert Text to Columns Wizard.
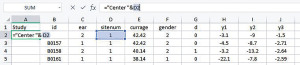
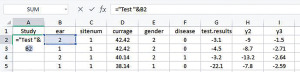
To facilitate the analyses provided in IPDmada, three columns are required and have to be named “Study”, “test.results”, and “disease”, and the variable names are case sensitive (e.g., Study must has a capital S). The column called “Study” indicates which primary study the records come from. In the analyses of a multiple centers study, we can use this column to reflect data are from which center. We insert one column to the left and set the header of this column as “Study”, and add a formula [=“Center”&D2] in cell A2 (Figure 3). If all the data are from one single center, just add the center name in cell A2.
Double click on the plus sign (+) in the bottom right corner of cell A2, then the formula (or value in case of single center study) is copied to all the rest cells in Column A.
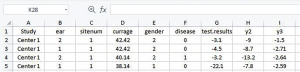
Change the header of Column G from “d” to “disease”, and change the header of Column H from “y1” to “test.results”. Remove Column B “id” which is not needed and may lead confusion to IPDmada (Figure 4).
After all these steps, data can be saved as a CSV file, and ready for uploading to IPDmada.
Analyses results
Upload data and check the correctness
Data will be shown in a table right after being uploaded (Figure 5). Please note that, because decimal point (.) and comma (,) are used in different ways in different regions in the world, the separator used in the CSV file may vary based on the system settings (regional settings in the control panel). If an error occurs, please switch the separator from “Semicolon” to “Comma” (or vice versa), usually this will solve the problem.
Calculation of test performance based on confusion matrix
Test performance calculated based on confusion matrix are shown in the “Analysis of dichotomized test results” panel.
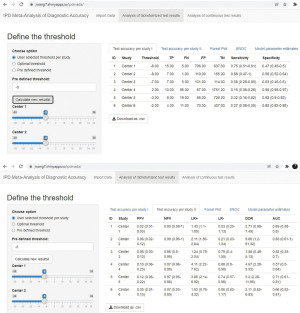
The threshold(s) can be determined by (I) entering different threshold values for each center with the slider bar; (II) using the optimal threshold values for each center; (III) entering one threshold value for all centers in the input box. The first option is useful when different thresholds are used in different centers, e.g., index test is from different manufacturers and the recommended thresholds are different; the second option is often desired when we need to determine an optimal threshold; the third option allows the users to tune the threshold for sensitivity analysis and data visualization purposes, to have a better understand how sensitivity and specificity will change when the threshold changes.
Numbers in the confusion matrix (i.e., TP, FN, FP, TN) and the corresponding test performance will be presented/updated after clicking the “Calculate new results!” button (Figure 6). Forest plots of sensitivity and specificity are also provided in the “Forest Plot” sheet (results not shown).
Calculation of test performance based on distribution of test results
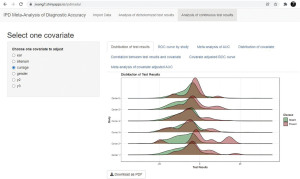
Before doing any calculation, the distribution of test results is visualized with a ridgeline plot (Figure 7). This ridgeline plot shows the distribution of index test in diseased group (Present) and non-diseased group (Absent), and smaller overlap between the two groups indicates better discrimination power of the index test.
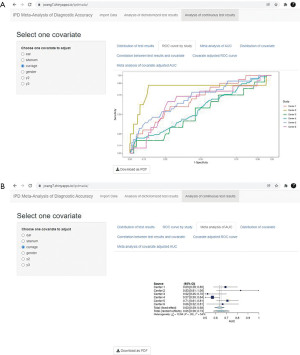
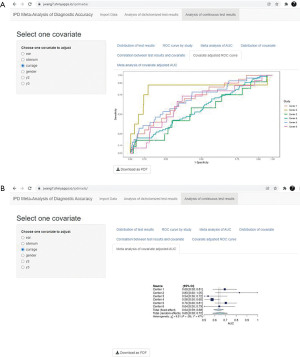
ROC curve of the test results is plotted for each center (Figure 8A), and the corresponding AUCs are summarized in a forest plot (Figure 8B). ROC curve shows the relation between TP rate/sensitivity (y-axis) and FP rate/(1−specificity) (x-axis) at all possible threshold values.
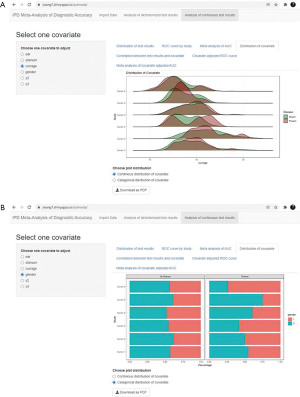
Advanced analyses of covariates are also provided (7). The distribution of a continuous covariate is plotted with a ridgeline plot (Figure 9A), while the distribution of a categorical covariate is plotted as a bar chart (Figure 9B). Investigating the distributions of covariates can help to identify differences in patient characteristics in diseased and non-diseased groups, which may influence the true test performance. If substantial differences are observed, we can consider using covariate adjusted ROC curve for further analysis.
In the end, covariate adjusted ROC curve of the test results is plotted for each center (Figure 10A), and the corresponding covariate AUCs are summarized in a forest plot (Figure 10B).
Analyses of data from a single center for multiple tests
Data preparation
If we want to analyze data of multiple tests from a single center, it can be done by changing the center name in Column “Study” to the test name.
We show this in a hypothetical example, where we treat tests performed on left ears and right ears as two different tests, and want to evaluate these two tests in one study. Please note that, the study design (i.e., paired data, unpaired data or randomized data) cannot be reflected in the analyses, and the numbers of patients underwent each test can be different. This is a limitation of using IPDmada in this situation: diagnostic accuracy performance measures can be calculated separately for each index test, however a formal comparison with statistical test between index tests is not supported.
We can start from the data shown in Figure 4. Add a formula [=“Test”&B2] in cell A2 (Figure 11), and double click on the plus sign (+) in the bottom right corner of cell A2 to copy the formula to all the rest cells in Column A (results not shown). After doing this, save the data as a CSV file.
Another example is, if the two (or more) tests being analyzed are saved in wide format (i.e., saved in two separate columns like y2 and y3), we need to transform the wide format data into long format and rename the column of test type as “Study”.
Analyses results
Analyses results are similar to those presented in multiple centers for one test, thus not shown again.
Discussion
IPDmada is a powerful tool, which can produce all analyses in a DTA study in a very efficient way. The time needed for data preparation and analyses is significantly reduced, and more importantly, less statistical knowledge is required from the researchers. All the analyses are facilitated by R, but the installation of R or running R locally is not needed. Compared to other statistical software, IPDmada is code-free and cost-free.
There are some limitations of IPDmada. First, the analyses provided by IPDmada are originally designed for IPD-MA, thus some of them may not be suitable for a primary DTA study. Second, when evaluating multiple index tests, the study design is not reflected and comparison between tests is not available. Third, IPDmada only provides standard analyses and has less flexibility. Last, all data preparation is done in a spreadsheet, which cannot be as convenient as statistical software.
In conclusion, IPDmada is an easy-to-use tool, which can provide a one-stop solution to researchers without strong statistical background for all the analyses needed in a DTA study.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Journal of Laboratory and Precision Medicine for the series “Statistics Corner”. The article has undergone external peer review.
Peer Review File: Avaible at https://jlpm.amegroups.com/article/view/10.21037/jlpm-22-13/prf
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://jlpm.amegroups.com/article/view/10.21037/jlpm-22-13/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bossuyt PM, Reitsma JB, Bruns DE, et al. The STARD statement for reporting studies of diagnostic accuracy: explanation and elaboration. Ann Intern Med 2003;138:W1-12. [Crossref] [PubMed]
- Shreffler J, Huecker MR. Diagnostic Testing Accuracy: Sensitivity, Specificity, Predictive Values and Likelihood Ratios 2022.
- Habbema JDF, Eijkemans R, Krijnen P, et al. Analysis of data on the accuracy of diagnostic tests. In: Knottnerus JA. The evidence base of clinical diagnosis. 1st edition. BMJ Books, 2002:117-44.
- Wang J, Keusters WR, Wen L, et al. IPDmada: An R Shiny tool for analyzing and visualizing individual patient data meta-analyses of diagnostic test accuracy. Res Synth Methods 2021;12:45-54. [Crossref] [PubMed]
- Norton SJ, Gorga MP, Widen JE, et al. Identification of neonatal hearing impairment: evaluation of transient evoked otoacoustic emission, distortion product otoacoustic emission, and auditory brain stem response test performance. Ear Hear 2000;21:508-28. [Crossref] [PubMed]
- Center FHCR. Diagnostic and Biomarkers Statistical (DABS) Center. Available online: https://research.fredhutch.org/diagnostic-biomarkers-center/en/datasets.html
- Janes H, Longton G, Pepe MS. Accommodating covariates in receiver operating characteristic analysis. Stata J 2009;9:17-39. [Crossref] [PubMed]
Cite this article as: Zhang X, Wang J. Using IPDmada to perform statistical analyses of diagnostic accuracy in primary studies: explanation and elaboration with a case study. J Lab Precis Med 2022;7:19.