
Unraveling the diagnostic enigma: laboratory diagnosis of sphingolipid activator protein deficiencies
Introduction
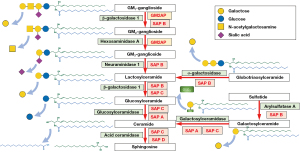
Sphingolipids, named after the mythical creature Sphinx due to their initially mysterious function akin to the enigmatic riddle posed by Sphinx, are essential components of the eukaryotic cell membrane (1). They are primarily localized in the nervous system and play a critical role in various cellular processes, such as regulating membrane dynamics, modulating cell signaling, and mediating cell adhesion and recognition (2). Based on the structure, sphingolipids can be categorized into sphingoid bases (and their simple derivatives), ceramides, and complex sphingolipids (3). Glycosphingolipids, a common type of complex sphingolipids, are characterized by the presence of carbohydrate groups in their molecules (4). Neutral glycosphingolipids, such as glucosylceramides (GlcCer) and galactosylceramides (GalCer), contain an uncharged carbohydrate group. Acidic glycosphingolipids, represented by sulfatides and gangliosides, comprise negatively charged groups such as sulfate groups or sialic acid groups (5) (Figure 1).

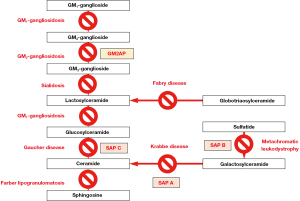
Sphingolipids are transported to lysosomes through the membrane flow, where they are degraded into the basic units (6). The degradation of sphingolipids is accomplished through a stepwise process involving various specific lysosomal acid hydrolases (7,8) (Figure 2). These enzymes can easily access glycosphingolipids with long-chain carbohydrate groups and efficiently break them down into smaller molecules. However, these enzymes cannot access glycosphingolipids with short oligosaccharide chains and require activator proteins to facilitate the degradation process (9). To date, five known sphingolipid activator proteins (SAPs) have been identified, including GM2 activator and four saposins (Saposin A, B, C, and D). SAPs exhibit membrane perturbing capabilities, act as physiological detergents, and/or engage in direct interactions with enzymes. These properties are indispensable for accessing and breaking down glycosphingolipids by lysosomal enzymes (10). Defects either in enzymes or SAPs lead to the inability to degrade certain sphingolipids. Consequently, these sphingolipids accumulate within lysosomes, causing sphingolipidoses, a subgroup of lysosomal storage disorders (11,12) (Figure 3).
The diagnosis of sphingolipidosis due to an enzyme deficiency can be accomplished by enzyme activity measurement (13). However, the diagnosis of SAP deficiencies remains challenging due to the complexities of SAP functional study in a clinical laboratory. Moreover, these disorders are exceptionally rare, increasing the likelihood of oversight and misdiagnosis. Because of the clinical similarities between SAP defects and corresponding enzyme deficiencies (14,15), physicians often encounter confusion caused by the discrepancy between clinical presentations and normal enzyme activities. This review paper focuses on laboratory methodologies and aims to provide insight into the enigmatic aspects surrounding the diagnosis of SAP deficiencies. By unraveling the complexities involved, it is hoped to streamline the laboratory diagnostic process, reducing diagnostic odysseys, and enhancing accuracy in identifying these defects.
GM2 activator deficiency
GM2 activator (GM2A or GM2AP), encoded by the gene GM2A, is a glycoprotein with a molecular weight of 17.6 kDa (16,17). GM2AP functions as the activator protein mainly for the degradation of GM2 gangliosides by hexosaminidase A (18). Its molecule possesses a hydrophobic cavity capable of harboring the ceramide moiety of GM2 gangliosides, which plays a vital role in delivering GM2 to hexosaminidase A (19). Any defect in hexosaminidase A or GM2AP results in the accumulation of GM2 gangliosides in lysosomes, causing GM2 gangliosidosis (19). Patients with GM2 gangliosidosis commonly present with impaired development of the central nervous system, exaggerated startle response, hypotonia, and cherry red spots (19). The majority of GM2 gangliosidosis cases are caused by hexosaminidase A deficiency, also known as Tay-Sachs disease (OMIM 272800), or combined hexosaminidase A and B deficiency, referred to as Sandhoff disease (OMIM 606873) (20). The diagnosis of Tay-Sachs disease or Sandhoff disease is established via enzyme assay, which reveals a significantly reduced activity (21). Sandhoff et al. observed an exceptional case of GM2 gangliosidosis with clinical features resembling those found in Tay-Sachs disease patients, yet hexosaminidases appeared to be normal when using a synthetic substrate in the enzyme assay (22,23). This exceptional case was subsequently confirmed as the first reported case of GM2 activator deficiency, also referred to as the AB variant of GM2 gangliosidosis (OMIM 272750) (24). Given that synthetic substrates are usually small molecules readily accessible and digestible by hexosaminidases, thus not requiring GM2AP, such substrates are unsuitable for assessing GM2 activator deficiency.
Functional study
O’Brien et al. demonstrated a significantly reduced hexosaminidase activity in fibroblasts from a patient with GM2 activator deficiency using natural GM2 ganglioside extracted from brain tissues as the substrate (25). It was evident that, in the absence of a functional GM2 activator, the natural GM2 gangliosides could not be degraded by the enzyme. Therefore, it is feasible to distinguish GM2 activator deficiency from other types of GM2 gangliosidoses by running parallel enzyme activity testing using synthetic and natural substrates respectively. In cases of GM2 activator deficiency, an abnormal activity result is obtained only when employing a natural substrate, while other types of GM2 gangliosidoses present abnormal results in both tests using synthetic and natural substrates. It is worth noting that, while the presence of detergent such as sodium taurocholate is crucial for achieving optimal enzyme activity (25,26), detergent quantity should be carefully validated since an excessive amount can compensate for the defect in GM2AP and result in false-negative results for GM2 activator deficiency. Tropak et al. established a fluorescence enzyme activity assay using a synthetic fluorescent GM2 analog that was incorporated into liposomes to mimic the intra-lysosomal membrane (27). By utilizing the modified liposomes along with recombinant GM2AP, they provided an alternative strategy for distinguishing GM2 activator deficiency from other GM2 gangliosidoses. That is, in GM2 activator deficiency, normal enzyme activity was observed in the presence of recombinant GM2AP, but abnormal activity was obtained when recombinant GM2AP was absent. In contrast, other GM2 gangliosidoses exhibited reduced enzyme activity regardless of the presence or absence of recombinant GM2AP (27). Several other techniques can be used to quantitively measure the expression of GM2AP, such as Western blotting, enzyme-linked immunosorbent assay (ELISA), screen-printed carbon electrode (SPCE), and fluorescence-resonance energy transfer (FRET) assay (28-30). Nonetheless, these techniques are solely suitable for situations where mutations in the GM2A gene result in decreased expression or absence of GM2AP, or critical alterations in protein antigenicity.
Given the complex nature of GM2AP functional studies, the measurement of biomarkers becomes essential during the diagnostic journey for GM2 activator deficiency. Abnormal levels of specific sphingolipid species along with typical clinical presentations in the setting of normal enzyme activity may strongly suggest an activator defect.
GM2 gangliosides
Initially, GM2 gangliosides were assessed using thin-layer chromatography (TLC), gas-liquid chromatography (GLC) and high-performance liquid chromatography (HPLC) (31-33). Tsuji et al. also established an ELISA method using an anti-GM2 monoclonal antibody to quantify GM2 in fibroblasts (34). The application of mass spectrometry (MS) for GM2 measurement offers an advantage due to its exceptional sensitivity. Tsui et al. developed a tandem mass spectrometry (MS/MS) strategy for the quantitative profiling of gangliosides in tissue lipid extracts, including GM2 (35). They employed an aqueous partition method for ganglioside enrichment and proposed a three-step MS analysis strategy, including precursor ion scan of 290 m/z, product ion spectrum analysis, and ceramide moiety composition determination (35). Subsequently, a simple and accurate isotope dilution method was also established to quantify GM2 human CSF samples with a linear range of 10–200 ng/mL (36). However, since GM2 gangliosides are integral components of cell membranes and are particularly abundant in brain tissue, the quantitative measurement of GM2 was mainly limited to cell or tissue extracts or CSF, significantly diminishing its clinical applicability (37,38). While an ultra-performance liquid chromatography (UPLC)-MS/MS method and an HPLC-MS/MS method were used to assess GM2 concentrations in the plasma of GM2 gangliosidosis patients, those studies suggested that GM2 species in plasma have limited diagnostic value (39,40). Blondel et al. validated an LC-MS/MS method to measure multiple plasma biomarkers following an easy sample preparation step (protein precipitation) (41). They discovered that GM2 (34:1) and the ratio of GM2 (34:1) to GM3 (34:1) were highly effective markers for distinguishing GM2 gangliosidoses from normal controls with 100% sensitivity and 100% specificity (41). Those markers were also able to differentiate GM2 gangliosidoses from other lysosomal storage disorders that may have secondary accumulation of gangliosides, including Niemann–Pick disease types A/B and C, GM1-gangliosidosis, Fabry disease, Gaucher disease and Krabbe disease (41). Remarkably, they also observed elevated levels of GM2 (34:1) in a patient with GM2 activator deficiency (41). Therefore, GM2 (34:1) potentially can be used to streamline the diagnosis of GM2 activator deficiency.
Other biomarkers
Welford et al. conducted an investigation into various biomarkers in plasma for gangliosidoses, including lysoGM2 and lysoGM1, neurofilament light chain (NF-L), and glial fibrillary acidic protein (GFAP) (42). LysoGM2 and lysoGM1 are deacylated derivatives of GM2 and GM1 gangliosides, respectively (43). NF-L is a neuron cytoskeletal protein and serves as a blood biomarker for neuronal damage (44). GFAP, on the other hand, is an astrocyte cytoskeletal filament as well as a biomarker for reactive astrocytosis (45). Using LC-MS/MS, Welford et al. observed elevated levels of lysoGM2 and lysoGM1 in almost all GM2 gangliosidosis patients, as well as in those with GM1 gangliosidosis (42). Additionally, through immunoassay, they also detected elevated NF-L and GFAP in those patients (42). However, although these biomarkers could be used for treatment monitoring, they are not sufficiently specific for the diagnosis of GM2 gangliosidosis (42). GM2 activator deficiency was not investigated in this study. With a multiplex LC-MS/MS, Pettazzoni et al. quantified several lysosphingolipids and found lysoGM2 was elevated in 10 out of 13 patients of Tay-Sachs and Sandhoff diseases but not detected in other diseases (46). Yet, lysoGM2 was reported to be normal in the plasma of a patient with a GM2 activator deficiency (47), indicating a limited clinical utility of lysoGM2.
Molecular genetic analysis of the GM2A gene is an indispensable step in ultimately diagnosing GM2 activator deficiency. To date, 26 cases of GM2 activator deficiency have been reported, ascribed to a range of identified genetic variants including missense, nonsense, frameshift mutations, and deletions (24,48-66).
Saposin A deficiency
Saposins are four small non-enzymatic proteins localized within lysosomes, namely saposin A, B, C, and D (67). All four saposins derive from a common precursor protein, referred to as prosaposin, which is encoded by the PASP gene (67). Prosaposin molecule contains four distinct functional domains (68). Upon transport to lysosomes, prosaposin undergoes hydrolysis, releasing these domains as individual saposin proteins (69). Saposin A functions as an activator protein for galactosylceramidase, facilitating the breakdown of GalCer to ceramides (70). Saposin A is also involved in the degradation of GlcCer by β-glucosidase; thus, its function partially overlaps with that of saposin C, although the role of saposin A in the degradation of GlcCer is to a lesser extent (70). The inability to degrade GalCer can result in Krabbe disease, which is typically caused by a deficiency in galactosylceramidase (OMIM 245200) (71). Saposin A deficiency (OMIM 611722) was first reported by Spiegel et al., who described a case of a six-month-old girl with atypical Krabbe disease characterized by progressive encephalopathy and abnormal myelination (72). The significantly decreased galactosylceramidase activity in the leukocytes of this girl, which was around 1% of the average of the normal range, initially led to a misdiagnosis of Krabbe disease due to galactosylceramidase deficiency (72). Nevertheless, they observed that the galactosylceramidase activity in fibroblasts was normalized in the presence of detergents, which suggested an activator deficiency (72). Saposin A deficiency was subsequently confirmed by identifying a homozygous mutation within the saposin A domain of the PSAP gene (72). Although the authors stated that the unexpected decrease in galactosylceramidase activity could be attributed to compromised sample integrity (72), the significantly reduced enzyme activity does not seem to be solely due to the non-fresh sample, as non-fresh samples typically exhibit mild to moderate reductions. In the second documented case of saposin A deficiency, Kose et al. also noticed that galactosylceramidase activities in both dried blood spots (DBS) and leukocytes were decreased, although the activity in leukocytes was above the affected range for typical Krabbe disease (73). In the third case of saposin A deficiency reported by Calderwood et al., galactosylceramidase enzyme activity in leukocytes was also slightly below the normal range but above the affected range (74). Those reports suggest that saposin A possibly plays a role in stabilizing the enzyme or increasing the maximal velocity of the enzyme (70). While galactosylceramidase activity in a freshly collected specimen might be less affected by saposin A defect, enzyme activity in a non-fresh specimen might be considerably compromised, significantly increasing the likelihood of being misdiagnosed as galactosylceramidase deficiency. While directly assessing saposin A functionality is currently not available in a clinical laboratory setting, quantification of saposin A amount in plasma can be achieved by time-delayed fluorescence immunoassays (75) or by immunoblotting (70). Nevertheless, these methods are only appropriate in the scenarios where genetic variants cause reduced or entirely absent protein expression or critical antigenicity alterations.
Due to the malfunction of saposin A, the impairment in the degradation of GalCer in skin fibroblasts was observed (72). Therefore, a prominent elevation of GalCer in fibroblast extractions was revealed using TLC, along with the elevations of GlcCer, lactosylceramides, and ceramides to a lesser extent (73). Considering the overlapping functionalities of saposin A and C but the relatively lesser role of saposin A in GlcCer degradation (70), GlcCer may lack distinctiveness as a biomarker. Moreover, an elevation of ceramides in saposin A deficiency became an enigmatic riddle, since as a product of GalCer and GlcCer degradation its elevation in saposin A deficiency is not anticipated. Unfortunately, this point was not discussed in the original report. Hence, more investigations were conducted on GalCer and its related metabolites as appropriate biomarkers for Krabbe disease, including saposin A deficiency.
GalCer
It was reported that GalCer primarily accumulated in localized globoid cells in the brain of Krabbe disease patients, but its total amount was generally low (76). While a slight elevation was detected in the kidney, no elevations were observed in the urine, CSF, and skin fibroblasts of Krabbe disease patients (77-79). To date, there have been no studies on GalCer levels in the blood of Krabbe disease patients that have yielded significant findings. Dawson reported a notable increase in the liver tissue, suggesting liver biopsy could be potentially valuable for diagnostic purposes (80). However, the invasive nature of biopsies has constrained the applicability of GalCer as a biomarker.
Psychosine
Psychosine, also known as galactosylsphingosine, is a cytotoxic byproduct resulting from GalCer deacylation and accumulates in the brain of Krabbe disease patients (81). The elevation of psychosine in the DBS from a saposin A deficiency patient was also reported (74). Several techniques were employed to measure psychosine levels in human and animal tissues, including TLC, HPLC, MS, and NMR spectroscopy (82-85). LC-MS and LC-MS/MS methods were also developed to quantify psychosine levels in mouse CSF and serum (86,87). Chuang et al. utilized HPLC-MS/MS to quantitatively detect psychosine in DBS from newborns and observed elevated psychosine levels in infantile patients with Krabbe disease (88). Subsequently, Turgeon et al. further validated this method and affirmed the significant potential of psychosine in DBS as a newborn screening biomarker (89). The role of psychosine in newborn screening for Krabbe disease was further confirmed through the analysis of longitudinal data and improved LC-MS/MS methods (90-92). LC-MS/MS methods for simultaneously quantifying psychosine and glucosylsphingosine (lyso-Gb1) in plasma, serum and CSF were also developed (93,94). It is worth noting that the lower limit of quantitation (LLOQ) for psychosine in CSF was 0.1 pg/mL, compared to the LLOQ of 0.03 ng/mL in plasma (94). It suggests that CSF has the potential to detect even minor fluctuations in psychosine levels, rendering it a more sensitive specimen type than plasma (94). Nonetheless, the invasive collection procedure makes CSF unsuitable for treatment monitoring. The utilization of psychosine in plasma and serum as a biomarker for diagnosing and monitoring Krabbe disease still requires further investigations.
When evaluating patients suspected of Krabbe disease, it is essential to proceed with caution to prevent misdiagnosis, as both galactosylceramidase and saposin A deficiencies can exhibit reduced galactosylceramidase activity. Particularly in cases where a molecular genetic analysis of GALC, the gene encoding galactosylceramidase, returns negative findings, the subsequent testing should involve molecular analysis of the PSAP gene. The diagnosis of saposin A deficiency is confirmed when molecular analysis reveals homozygous or compound heterozygous pathogenic variants within the saposin A domain of the PSAP gene. Alternatively, the presence of one allele carrying a heterozygous pathogenic variant within the saposin A domain, along with another deleterious allele affecting the entire prosaposin protein, can also establish the diagnosis. In the three reported patients, homozygous pathogenic or likely pathogenic variants in the coding region of the saposin A domain, including the single base pair deletion c.207_209del, the missense variant c.209T>G, and the missense variant c.257T>A, were identified (72-74).
Saposin B deficiency
Saposin B was initially discovered by Mehl et al. and demonstrates broad-ranging specificity (10,95). It can extract sphingolipids of multiple types from the membrane and deliver them to their respective enzymes for degradation (10). Saposin B primarily stimulates the hydrolysis of GM1 gangliosides, sulfatides, and globotriaosylceramide (Gb3) (96-98). Sulfatides are digested by arylsulfatase A, the deficiency of which causes metachromatic leukodystrophy (MLD) (OMIM 250100) (99). Gb3 is a substrate for α-galactosidase, a defect which leads to Fabry disease (OMIM 301500) (100). GM1 ganglioside degradation relies on β-galactosidase 1, and its deficiency underlies GM1-gangliosidosis (OMIM 230500, 230600, 230650) and mucopolysaccharidosis type IVB (OMIM 253010) (101). The accumulation of these sphingolipids and their derivatives concurrently would strongly indicate saposin B deficiency (OMIM 249900). Shapiro et al. reported that two siblings with characteristic symptoms of MLD exhibited a defect in sulfatide degradation, despite having nearly normal arylsulfatase A activity (102). Subsequent studies confirmed that those siblings were indeed affected by saposin B deficiency (103,104). Fujibayashi et al. developed a simple immunoblotting method using an anti-saposin B antibody to screen for saposin B deficiency (105). This method applies exclusively to patients with deleterious mutations that lead to reduced protein expression or critical antigenicity alterations. Fischer et al. set up an enzyme assay using purified arylsulfatase A and radiolabeled sulfatide to evaluate the activating effect of saposin B (106). This assay relies on the principle that the degradation of sulfatides by the purified arylsulfatase A requires the presence of either a detergent like taurodeoxycholate or functional saposin B protein (106). To eliminate the use of hazardous radioactive materials and laborious preparation procedure, Norris et al. also developed an ESI-MS/MS method to quantify saposin B activity based on the same strategy and demonstrated this nonradioactive method was reliable, fast and specific (107). These initiatives have made it possible to measure saposin B activity in human specimens for the diagnosis of saposin B deficiency. However, there have been no reports of establishing such a diagnostic test in a clinical laboratory setting yet.
Sulfatides
Urinary excretion of sulfatides is a characteristic biochemical feature of MLD (108). Sulfatides can be measured by TLC, spectrophotometry, HPTLC, or HPLC methods (109-112). Hsu et al. developed an ESI-MS/MS method to measure sulfatides in brain and pancreatic tissues (113). Whitfield et al. expanded an MS/MS method for urine sulfatide measurement and detected substantially increased levels of multiple hydroxy and non-hydroxy species of sulfatides in the urine samples from MLD patients (114). Cui et al. made further enhancements to the MS/MS method by synthesizing a new non-physiological sulfatide as an internal standard (115). Through MS/MS coupled with solid-phase extraction, Kuchař et al. identified five sulfatide isoforms, namely C22:0, C22:0-OH, C24:0, C24:1-OH, and C24:0-OH sulfatides, which exhibited the most significant distinctions between urine samples from MLD patients and the control group (116). LC-MS/MS methods on DBS and dried urine spots (DUS) were also established (117,118). Barcenas et al. demonstrated that the sulfatide levels in the DUS from MLD patients did not overlap with those from non-MLD patients, whereas such an overlap existed in the DBS from MLD and non-MLD patients, indicating that DUS is a better specimen type compared to DBS (118).
Lysosulfatides
Lysosulfatides, the deacylation product of sulfatides, were detected by HPLC and reported to be elevated in the tissues of MLD patients (119). Several methods based on matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) MS methods were also developed to measure lysosulfatides (120-122). Mirzaian et al. established a LC-MS/MS method, by which they could quantify a wide range of sulfatide and lysosulfatide species in urine and plasma (123). Their findings suggested that sulfatides in plasma might lack sufficient sensitivity to reliably distinguish all MLD patients from healthy individuals, and that lysosulfatides might not be reliably quantifiable due to a potential interfering ion from sulfatides (123). Cao et al. also developed an LC-MS/MS method to quantify sulfatides and lysosulfatides in CSF (124). While there are various specimen types available for MLD biomarker measurement, urine sulfatides measurement, instead of lysosulfatides, likely remains the most clinically valuable test.
Gb3
As a biomarker for the X-linked disorder Fabry disease (OMIM 301500) (125), Gb3 is anticipated to also serve as a potential discriminative marker for distinguishing saposin B deficiency from arylsulfatase A deficiency. Methods such as TLC, gas chromatography (GC), and HPLC were initially developed to measure Gb3 at an earlier time (126-128). Zeidner et al. established a rapid and sensitive ELISA method to quantify Gb3 levels in plasma using an antibody with a distinct affinity for Gb3 (129). MS/MS method was also established by Boscaro and collaborators to measure Gb3 in human plasma and urine (130). Auray-Blais et al. utilized LC-MS/MS to quantify Gb3 on filter paper and found that it remained stable for 7 weeks in DBS (131). A new strategy of internal standard synthesis potentially expanded the capacity to measure various forms of Gb3 through MS (132). While Gb3 can be reliably measured in plasma and urine, Young et al.’s study revealed that plasma Gb3 levels were elevated exclusively in male patients with the classic form of Fabry disease, and the levels exhibited a wide range overlapping with those in the normal control group (133). Therefore, plasma Gb3 was not deemed an ideal marker for Fabry disease and urine Gb3 may possess greater diagnostic potential as a biomarker (133). Kuchař et al. reported that urinary Gb3 was significantly elevated in a saposin B deficiency patient, in addition to elevations of sulfatides (134).
Lyso-Gb3 and methylated Gb3
Lyso-Gb3 is a deacylation product of Gb3 by acid ceramidase (135). Togawa et al. reported that plasma lyso-Gb3 levels were elevated in both male and female Fabry patients, whereas plasma Gb3 levels in male patients with variant forms of Fabry disease and female patients were indistinguishable from those in the control group, suggesting that lyso-Gb3 may serve as a better biomarker compared to Gb3 (136). Auray-Blais et al.’s study demonstrated that urinary lyso-Gb3 is a reliable independent biomarker for Fabry disease (137). With UPLC-MS/MS, Gold et al. reported that both lyso-Gb3 and lyso-ene-Gb3 can be reliable biomarkers (138). Lavoie et al. developed a multiplex LC-MS/MS method to analyze lyso-Gb3 analogs in urine and identified the most abundant analogs with m/z 802, 820, and 836 as promising urinary biomarkers for Fabry disease (139). Metabolomic profiling with a time-of-flight (TOF)-MS unveiled more potential novel biomarkers for Fabry disease, including methylated Gb3-related isoforms (140). Abaoui et al.’s study suggested that methylated Gb3 isoforms are the intermediate metabolites between Gb3 and lyso-Gb3, thereby expanding the biomarker panel for Fabry disease (141). Regarding saposin B deficiency, further investigations into lyso-Gb3 are necessary to assess its potential clinical utility as a biomarker.
GM1 gangliosides
It has been reported that saposin B deficiency does not lead to the accumulation of keratan sulfate, which is a hallmark of mucopolysaccharidosis type IVB (142). Since saposin B is necessary for the degradation of GM1 gangliosides by β-galactosidase 1, it was hypothesized that saposin deficiency may result in the accumulation of GM1 gangliosides (143,144). However, only a slight elevation of GM1 gangliosides was observed in the skin fibroblasts from a patient with saposin B deficiency, while a higher elevation of GM3 gangliosides was detected instead (145). Due to limited research on GM1 gangliosides and the related metabolites as biomarkers for saposin B deficiency, it is not recommended to include them in clinical diagnostic testing currently.
Saposin B deficiency is the most common sphingolipid activator deficiency, with more than 30 cases have been reported (146-148). The diagnosis of saposin B deficiency is established through molecular genetic analysis of the PSAP gene, which identifies either pathogenic variants within the saposin B domain on each allele, or one allele with a pathogenic variant in the saposin B domain and another allele with a pathogenic variant located outside of saposin B domain but damaging the entire prosaposin protein.
Saposin C deficiency
Saposin C protein was first isolated by Ho et al. (149). Similar to saposin B, saposin C also exhibits a broad specificity. Saposin C can stimulate the degradation of GlcCer by acid β-glucosidase, the degradation of GalCer by galactosylceramidase, and the degradation of sphingomyelins by sphingomyelinase (150-152). Saposin C deficiency (OMIM 610539) was initially reported by Christomanou et al., who demonstrated that a variant form of Gauche disease presented by a patient stemmed from the absence of Saposin C (153). In contrast to typical Gaucher disease (OMIM 230800, 230900, 231000) caused by acid β-glucosidase deficiency, patients with saposin C deficiency exhibited a normal β-glucosidase activity when a synthetic substrate such as 4-methylumbelliferyl-β-D-glucopyranoside was used for enzyme assay (154,155). When a natural substrate such as GlcCer was used, saposin C deficiency patients had reduced β-glucosidase activity (153). Chang et al. introduced a fluorescence immunoassay for quantifying saposin C concentration in plasma (75). Meikle et al. utilized this method to assess the amount of saposin C protein in DBS (156). Those methods are convenient in cases where saposin C deficiency is due to mutations leading to reduced expression or the absence of saposin C or antigenicity alterations. Yoneshige et al. directly quantified the activity of synthetic saposin C using imiglucerase (Cerezyme®), which is a recombinant analogue of β-glucosidase, and artificial fluorescent substrates embedded into the liposome to mimic intra-lysosomal membrane (157). This strategy can be employed by a clinical lab to determine saposin C activity in patient samples. Nonetheless, it’s worth noting that the clinical use of this strategy as a diagnostic tool for saposin C deficiency has not been reported yet.
A variety of biomarkers have been discovered for Gaucher disease, including tartrate-resistant acid phosphatase (TRAP), angiotensin-converting enzyme (ACE), cathepsin K, Chitotriosidase, pulmonary and activation-regulated chemokine (PARC/CCL18), chemokines macrophage inflammatory protein (MIP)-1α and MIP-1β, ferritin, GlcCer, GM3 gangliosides and lyso-Gb1 (158,159). Elevations of chitotriosidase activity, chemokine CCL18, GlcCer, and lyso-Gb1 were also observed in the plasma of patients with saposin C deficiency (154,159,160). Since many of these biomarkers are associated with secondary abnormalities and lack sufficient specificity and sensitivity (158), only GlcCer and its related metabolites are discussed in the following section.
GlcCer
GlcCer was initially detected using methods such as GC and HPLC (161-163). Samuelsson et al. developed a GC-MS method for GlcCer detection (164). Murata et al. also reported a direct inlet-chemical ionization MS that was better for the determination and structural studies of cerebroside species including GlcCer (165). Hillborg et al. first reported the elevation of GlcCer in plasma from Gaucher patients (166). Multiple studies further confirmed that its plasma levels were elevated in Gaucher disease (161,162). Through an ESI-MS/MS method, Whitfield et al. observed a general increase in all species of GlcCer in the plasma of Gaucher patients; however, there was a substantial overlap between the levels detected in the affected group and those in the normal group (167). They also noticed that the ratio of GlcCer (C16:0)/lactosylceramide (C16:0) was approximately four times higher in the patient than in normal individuals, indicating its potential as a valuable diagnostic biomarker (167). When exploring newborn screening biomarkers with LC-MS/MS, Meikle et al. also reported that GlcCer (C16:0) was elevated only in 3 of 5 Gaucher patients with sensitivity and specificity of 60 and 100, respectively (156). Therefore, those studies suggest that the ratio of GlcCer (C16:0) to lactosylceramide (C16:0), instead of GlcCer (C16:0) alone, holds the potential of greater clinical value and is worthy of further investigations.
Lyso-Gb1
Lyso-Gb1 is a product of GlcCer deacylation by acid ceramidase (135). The elevations of lyso-Gb1 in the spleen, hepatic, and brain tissues of Gaucher patients, as well as in human fetuses affected with type 2 Gaucher disease, were reported (168-171). In those studies, the detection methods included TLC, GLC, or HPLC (168-170). Oshima reported using a GC-MS method to identify lyso-Gb1 in the spleen tissue of Gaucher patients (172). An HPLC-FAB-MS method was also established, which enabled accurate quantification of lyso-Gb1 due to its ability to effectively separate lyso-Gb1 from psychosine (173). Dekker et al. utilized an LC-ESI-MS/MS technique to quantify lyso-Gb1 and revealed a significant elevation of more than 200 folds in the plasma of Gaucher patients, without an overlap with the control group (159). Comparable elevations of lyso-Gb1 were also observed in the plasma of two patients with saposin C deficiency (159). Rolfs et al. also detected increased lyso-Gb1 levels in the plasma of patients with Gaucher disease using HPLC-MS/MS and determined an optimal cut-off of 12 ng/mL with both sensitivity and specificity of 100% (174). Their study provides strong evidence that lyso-Gb1 is an exceptionally sensitive and specific biomarker for Gaucher disease (174). Mirzaian et al. optimized the LC-ESI-MS/MS method by incorporating an isotope-labelled lyso-Gb1 standard to improve test sensitivity and enable the measurement of lyso-Gb1 in unconcentrated urine samples (175). They further demonstrated elevated lyso-Gb1 levels in the urine of symptomatic type 1 GD patients (175). Zhang et al. developed a hydrophilic interaction liquid chromatography (HILIC)-MS/MS method to reliably detect lyso-Gb1 in DBS, thus streamlining the utilization of lyso-Gb1 in newborn screening as a biomarker for Gaucher disease (176). Quantification of lyso-Gb1 in CSF was also reported (94). Hence, lyso-Gb1 stands as a highly sensitive and convenient biomarker for the diagnosis of Gaucher disease, as well as saposin C deficiency, as systematically reviewed by Revel-Vilk et al. (177). Moreover, metabolomics studies have unveiled related lyso-Gb1 analogs as potential biomarkers (178). Subsequent investigations have explored their role in the diagnosis of Gaucher disease (179-181). Nevertheless, those analogs will require further assessments in future research.
To date, 15 cases of saposin C deficiency have been reported (14,153-155,160,182-184). The diagnosis of saposin C deficiency is confirmed through molecular analysis of the PSAP gene, which reveals the presence of two pathogenic variants in-trans, with at least one affecting the saposin C domain, while the other impacting either saposin C or the entire protein.
Saposin D deficiency
Saposin D, as an activator for sphingomyelinase and ceramidase, facilitates the breakdown of sphingomyelins and ceramides (185,186). Matsuda et al. reported that a mouse model of saposin D deficiency presented with the accumulation of ceramides in the brain tissues (187). However, saposin D deficiency has not been reported in humans, potentially due to the broad specificity of other saposin proteins, particularly saposin C, which can compensate for the dysfunction of saposin D in the degradation of sphingomyelins and ceramides (150,188). Although it has been reported that pathogenic variants within the saposin D domain of the PSAP gene are associated with Parkinson’s disease (189), this discovery remains a topic of controversy (190,191).
Prosaposin deficiency
Prosaposin deficiency (OMIM 611721), also known as combined saposin deficiencies, is attributed to the impairment of the entire prosaposin protein. Harzer et al. reported two siblings presented with an atypical form of Gaucher disease that was characterized by more severe symptoms, the accumulation of GlcCer and ceramides in liver tissue, as well as the partially reduced activities of galactosylceramidase, β-glucosidase, and ceramidase (192). Those two siblings possessed homozygous mutations in the initiation codon of the PSAP gene, which could have led to either the absence of prosaposin protein or mistargeting of prosaposin into medium instead of transport to lysosomes (193). Additional biochemical findings in those siblings included the absence of both saposin B and C in lysosomes, as well as the elevations of various neutral glycolipids in multiple tissues, including mono-, di-, tri-, and tetrahexosylceramides such as GlcCer, GalCer, lactosylceramides, and digalactosylceramides, in addition to sulfatides, ceramides, GM3 and GM2 gangliosides (194,195).
There have been 10 reported cases of prosaposin deficiency (134,192,196-200). Summarizing those ten cases reveals that patients with prosaposin deficiency typically present with more severe clinical symptoms, including hepatosplenomegaly, microcephaly, hypotonia, seizures, optic atrophy, and other manifestations (198,199). Multiple types of sphingolipids accumulate in various tissues, including lyso-Gb1, lyso-Gb3, psychosine, ceramides, GlcCer, lactosylceramides, GalCer, Gb3, sulfatides and GM3 gangliosides (198,199). Therefore, quantitative analysis of plasma lysosphingolipids by LC-MS/MS demonstrated a specific pattern of combined saposin deficiency, particularly elevations of both lyso-Gb1 and lyso-Gb3 (199). The enzyme assays revealed varying degrees of reduced activities in galactosylceramidase, β-glucosidase and ceramidase in leukocytes or fibroblasts, whereas the activities of other enzymes such as sphingomyelinase and neuraminidase remained within the normal range (192,199). Hence, a plasma or urine lysosphingolipid panel test that incorporates lyso-Gb1 and lyso-Gb3 could serve as a valuable tool for the early diagnosis of prosaposin deficiency. The diagnosis is confirmed through molecular genetic analysis, which identifies two in-trans pathogenic variants resulting in the malfunction of the entire prosaposin protein (199). The reported pathogenic variants caused frameshift, premature stop, or a splicing error in the PSAP gene, leading to the absence or loss of function of prosaposin protein and affecting multiple saposin proteins (134,198,199). Similar to saposin A deficiency, it is crucial to proceed with caution when discrepancies exist between reduced enzyme activities and negative molecular genetic analysis of the relevant enzyme genes. This is especially important when elevated levels of multiple biomarkers associated with different activator proteins are detected. In such cases, it is appropriate to pursue molecular analysis of the PSAP gene to confirm the diagnosis of prosaposin deficiency.
Conclusions
Lysosomal degradation of glycosphingolipids relies on the presence of SAPs. It is challenging to diagnose SAP deficiencies as affected individuals usually exhibit normal enzyme activities despite presenting with typical clinical features. Biomarkers such as GM2 ganglioside (34:1), sulfatides, lyso-GB1, lyso-Gb3, and psychosine are valuable for addressing this diagnostic complexity, and molecular genetics analysis serves as the essential confirmatory test. In cases of saposin A and prosaposin deficiencies, it’s noteworthy that abnormal enzyme activities can be observed. This could potentially result in the misdiagnosis of corresponding enzyme deficiencies. In such scenarios, discrepancies between reduced enzyme activities and negative findings in molecular genetic analysis of the relevant enzyme genes may indicate the necessity of a follow-up molecular test targeting the PSAP gene.
Ongoing research in this field may further advance our understanding of SAP deficiencies and improve the current diagnostic and therapeutic approaches.
Acknowledgments
Funding: None.
Footnote
Peer Review File: Available at https://jlpm.amegroups.org/article/view/10.21037/jlpm-23-71/prf
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at https://jlpm.amegroups.org/article/view/10.21037/jlpm-23-71/coif). The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- De Nicolò A, Cantù M, D'Avolio A. Matrix effect management in liquid chromatography mass spectrometry: the internal standard normalized matrix effect. Bioanalysis 2017;9:1093-105. [Crossref] [PubMed]
- Posse de Chaves E, Sipione S. Sphingolipids and gangliosides of the nervous system in membrane function and dysfunction. FEBS Lett 2010;584:1748-59. [Crossref] [PubMed]
- Quinville BM, Deschenes NM, Ryckman AE, et al. A Comprehensive Review: Sphingolipid Metabolism and Implications of Disruption in Sphingolipid Homeostasis. Int J Mol Sci 2021;22:5793. [Crossref] [PubMed]
- Vakirtzi-Lemonias C, Evangelatos GP, Kapoulas VM, et al. Studies on glycosphingolipid biosynthesis by lectin-stimulated human lymphocytes. Eur J Biochem 1980;109:541-51. [Crossref] [PubMed]
- Ryckman AE, Brockhausen I, Walia JS. Metabolism of Glycosphingolipids and Their Role in the Pathophysiology of Lysosomal Storage Disorders. Int J Mol Sci 2020;21:6881. [Crossref] [PubMed]
- Kok JW, Babia T, Hoekstra D. Sorting of sphingolipids in the endocytic pathway of HT29 cells. J Cell Biol 1991;114:231-9. [Crossref] [PubMed]
- Schwarzmann G, Sandhoff K. Metabolism and intracellular transport of glycosphingolipids. Biochemistry 1990;29:10865-71. [Crossref] [PubMed]
- Schuette CG, Doering T, Kolter T, et al. The glycosphingolipidoses-from disease to basic principles of metabolism. Biol Chem 1999;380:759-66. [Crossref] [PubMed]
- Schuette CG, Pierstorff B, Huettler S, et al. Sphingolipid activator proteins: proteins with complex functions in lipid degradation and skin biogenesis. Glycobiology 2001;11:81R-90R. [Crossref] [PubMed]
- Fürst W, Sandhoff K. Activator proteins and topology of lysosomal sphingolipid catabolism. Biochim Biophys Acta 1992;1126:1-16. [Crossref] [PubMed]
- Wenger DA, Luzi P, Rafi MA. Lysosomal storage diseases: heterogeneous group of disorders. Bioimpacts 2013;3:145-7. [PubMed]
- Schuchman EH, Simonaro CM. The genetics of sphingolipid hydrolases and sphingolipid storage diseases. Handb Exp Pharmacol 2013;3-32. [Crossref] [PubMed]
- Bekri S. Laboratory diagnosis of lysosomal storage diseases. In: Mehta A, Beck M, Sunder-Plassmann G, editors. Fabry Disease: Perspectives from 5 Years of FOS. Oxford: Oxford PharmaGenesis; 2006.
- Kang L, Zhan X, Ye J, et al. A rare form of Gaucher disease resulting from saposin C deficiency. Blood Cells Mol Dis 2018;68:60-5. [Crossref] [PubMed]
- Madaan P, Jauhari P, Chakrabarty B, et al. Saposin B-Deficient Metachromatic Leukodystrophy Mimicking Acute Flaccid Paralysis. Neuropediatrics 2019;50:318-21. [Crossref] [PubMed]
- Burg J, Conzelmann E, Sandhoff K, et al. Mapping of the gene coding for the human GM2 activator protein to chromosome 5. Ann Hum Genet 1985;49:41-5. [Crossref] [PubMed]
- Wright CS, Zhao Q, Rastinejad F. Structural analysis of lipid complexes of GM2-activator protein. J Mol Biol 2003;331:951-64. [Crossref] [PubMed]
- Kolter T, Sandhoff K. Principles of lysosomal membrane digestion: stimulation of sphingolipid degradation by sphingolipid activator proteins and anionic lysosomal lipids. Annu Rev Cell Dev Biol 2005;21:81-103. [Crossref] [PubMed]
- Leal AF, Benincore-Flórez E, Solano-Galarza D, et al. GM2 Gangliosidoses: Clinical Features, Pathophysiological Aspects, and Current Therapies. Int J Mol Sci 2020;21:6213. [Crossref] [PubMed]
- Gilbert F, Kucherlapati R, Creagan RP, et al. Tay-Sachs' and Sandhoff's diseases: the assignment of genes for hexosaminidase A and B to individual human chromosomes. Proc Natl Acad Sci U S A 1975;72:263-7. [Crossref] [PubMed]
- Wendeler M, Sandhoff K. Hexosaminidase assays. Glycoconj J 2009;26:945-52. [Crossref] [PubMed]
- Sandhoff K, Andreae U, Jatzkewitz H. Deficient hexosaminidase activity in an exceptional case of Tay-Sachs disease with additional storage of kidney globoside in visceral organs. Pathol Eur 1968;3:278-85. [Crossref] [PubMed]
- Sandhoff K. Variation of beta-N-acetylhexosaminidase-pattern in Tay-Sachs disease. FEBS Lett 1969;4:351-4. [Crossref] [PubMed]
- Sandhoff K, Harzer K, Wässle W, et al. Enzyme alterations and lipid storage in three variants of Tay-Sachs disease. J Neurochem 1971;18:2469-89. [Crossref] [PubMed]
- O'Brien JS, Norden GW, Miller AL, et al. Ganglioside GM2 N-acetyl-beta-D-galactosaminidase and asialo GM2 (GA2) N-acetyl-beta-D-galactosaminidase; studies in human skin fibroblasts. Clin Genet 1977;11:171-83. [Crossref] [PubMed]
- Svennerholm L, Håkansson G, Månsson JE, et al. The assay of sphingolipid hydrolases in white blood cells with labelled natural substrates. Clin Chim Acta 1979;92:53-64. [Crossref] [PubMed]
- Tropak MB, Bukovac SW, Rigat BA, et al. A sensitive fluorescence-based assay for monitoring GM2 ganglioside hydrolysis in live patient cells and their lysates. Glycobiology 2010;20:356-65. [Crossref] [PubMed]
- Potprommanee L, Ma HT, Shank L, et al. GM2-activator protein: a new biomarker for lung cancer. J Thorac Oncol 2015;10:102-9. [Crossref] [PubMed]
- Kuntamung K, Sangthong P, Jakmunee J, et al. A label-free immunosensor for the detection of a new lung cancer biomarker, GM2 activator protein, using a phosphomolybdic acid/polyethyleneimine coated gold nanoparticle composite. Analyst 2021;146:2203-11. [Crossref] [PubMed]
- Schwarzmann G, Wendeler M, Sandhoff K. Synthesis of novel NBD-GM1 and NBD-GM2 for the transfer activity of GM2-activator protein by a FRET-based assay system. Glycobiology 2005;15:1302-11. [Crossref] [PubMed]
- Hoffman LM, Amsterdam D, Schneck L. GM2 ganglioside in fetal Tay-Sachs disease brain cultures: a model system for the disease. Brain Res 1976;111:109-17. [Crossref] [PubMed]
- Bremer EG, Gross SK, McCluer RH. Quantitative analysis of monosialogangliosides by high-performance liquid chromatography of their perbenzoyl derivatives. J Lipid Res 1979;20:1028-35. [Crossref] [PubMed]
- Lee WM, Westrick MA, Macher BA. High-performance liquid chromatography of long-chain neutral glycosphingolipids and gangliosides. Biochim Biophys Acta 1982;712:498-504. [Crossref] [PubMed]
- Tsuji D, Higashine Y, Matsuoka K, et al. Therapeutic evaluation of GM2 gangliosidoses by ELISA using anti-GM2 ganglioside antibodies. Clin Chim Acta 2007;378:38-41. [Crossref] [PubMed]
- Tsui ZC, Chen QR, Thomas MJ, et al. A method for profiling gangliosides in animal tissues using electrospray ionization-tandem mass spectrometry. Anal Biochem 2005;341:251-8. [Crossref] [PubMed]
- Gu J, Tifft CJ, Soldin SJ. Simultaneous quantification of GM1 and GM2 gangliosides by isotope dilution tandem mass spectrometry. Clin Biochem 2008;41:413-7. [Crossref] [PubMed]
- Ohmi Y, Ohkawa Y, Yamauchi Y, et al. Essential roles of gangliosides in the formation and maintenance of membrane microdomains in brain tissues. Neurochem Res 2012;37:1185-91. [Crossref] [PubMed]
- Bernheimer H, Molzer B, Deisenhammer E. Sandhoff disease: ganglioside G(M2) and asialo-G(M2) accumulation in the cerebrospinal fluid. J Neurochem 1977;29:351-3. [Crossref] [PubMed]
- Huang Q, Zhou X, Liu D, et al. A new liquid chromatography/tandem mass spectrometry method for quantification of gangliosides in human plasma. Anal Biochem 2014;455:26-34. [Crossref] [PubMed]
- Fuller M, Duplock S, Hein LK, et al. Liquid chromatography/electrospray ionisation-tandem mass spectrometry quantification of GM2 gangliosides in human peripheral cells and plasma. Anal Biochem 2014;458:20-6. [Crossref] [PubMed]
- Blondel A, Kraoua I, Marcelino C, et al. Plasma G(M2) ganglioside potential biomarker for diagnosis, prognosis and disease monitoring of GM2-Gangliosidosis. Mol Genet Metab 2023;138:106983. [Crossref] [PubMed]
- Welford RWD, Farine H, Steiner M, et al. Plasma neurofilament light, glial fibrillary acidic protein and lysosphingolipid biomarkers for pharmacodynamics and disease monitoring of GM2 and GM1 gangliosidoses patients. Mol Genet Metab Rep 2022;30:100843. [Crossref] [PubMed]
- Neuenhofer S, Conzelmann E, Schwarzmann G, et al. Occurrence of lysoganglioside lyso-GM2 (II3-Neu5Ac-gangliotriaosylsphingosine) in GM2 gangliosidosis brain. Biol Chem Hoppe Seyler 1986;367:241-4. [Crossref] [PubMed]
- Bischof A, Manigold T, Barro C, et al. Serum neurofilament light chain: a biomarker of neuronal injury in vasculitic neuropathy. Ann Rheum Dis 2018;77:1093-4. [Crossref] [PubMed]
- Yang Z, Wang KK. Glial fibrillary acidic protein: from intermediate filament assembly and gliosis to neurobiomarker. Trends Neurosci 2015;38:364-74. [Crossref] [PubMed]
- Pettazzoni M, Froissart R, Pagan C, et al. LC-MS/MS multiplex analysis of lysosphingolipids in plasma and amniotic fluid: A novel tool for the screening of sphingolipidoses and Niemann-Pick type C disease. PLoS One 2017;12:e0181700. [Crossref] [PubMed]
- Kodama T, Togawa T, Tsukimura T, et al. Lyso-GM2 ganglioside: a possible biomarker of Tay-Sachs disease and Sandhoff disease. PLoS One 2011;6:e29074. [Crossref] [PubMed]
- Hechtman P, Gordon BA, Ng Ying Kin NM. Deficiency of the hexosaminidase A activator protein in a case of GM2 gangliosidosis; variant AB. Pediatr Res 1982;16:217-22. [Crossref] [PubMed]
- de Baecque CM, Suzuki K, Rapin I, et al. GM2-gangliosidosis, AB variant: clinico-pathological study of a case. Acta Neuropathol 1975;33:207-26. [Crossref] [PubMed]
- Chen B, Rigat B, Curry C, et al. Structure of the GM2A gene: identification of an exon 2 nonsense mutation and a naturally occurring transcript with an in-frame deletion of exon 2. Am J Hum Genet 1999;65:77-87. [Crossref] [PubMed]
- Schepers U, Glombitza G, Lemm T, et al. Molecular analysis of a GM2-activator deficiency in two patients with GM2-gangliosidosis AB variant. Am J Hum Genet 1996;59:1048-56. [PubMed]
- Schröder M, Schnabel D, Hurwitz R, et al. Molecular genetics of GM2-gangliosidosis AB variant: a novel mutation and expression in BHK cells. Hum Genet 1993;92:437-40. [Crossref] [PubMed]
- Fernández-Marmiesse A, Morey M, Pineda M, et al. Assessment of a targeted resequencing assay as a support tool in the diagnosis of lysosomal storage disorders. Orphanet J Rare Dis 2014;9:59. [Crossref] [PubMed]
- Smith NJ, Winstone AM, Stellitano L, et al. GM2 gangliosidosis in a UK study of children with progressive neurodegeneration: 73 cases reviewed. Dev Med Child Neurol 2012;54:176-82. [Crossref] [PubMed]
- Sakuraba H, Itoh K, Shimmoto M, et al. GM2 gangliosidosis AB variant: clinical and biochemical studies of a Japanese patient. Neurology 1999;52:372-7. [Crossref] [PubMed]
- Sheth J, Datar C, Mistri M, et al. GM2 gangliosidosis AB variant: novel mutation from India - a case report with a review. BMC Pediatr 2016;16:88. [Crossref] [PubMed]
- Salih MA, Seidahmed MZ, El Khashab HY, et al. Mutation in GM2A Leads to a Progressive Chorea-dementia Syndrome. Tremor Other Hyperkinet Mov (N Y) 2015;5:306. [Crossref] [PubMed]
- Renaud D, Brodsky M. GM2-Gangliosidosis, AB Variant: Clinical, Ophthalmological, MRI, and Molecular Findings. JIMD Rep 2016;25:83-6. [Crossref] [PubMed]
- Hall PL, Laine R, Alexander JJ, et al. GM2 Activator Deficiency Caused by a Homozygous Exon 2 Deletion in GM2A. JIMD Rep 2018;38:61-5. [Crossref] [PubMed]
- Martins C, Brunel-Guitton C, Lortie A, et al. Atypical juvenile presentation of GM2 gangliosidosis AB in a patient compound-heterozygote for c.259G > T and c.164C > T mutations in the GM2A gene. Mol Genet Metab Rep 2017;11:24-9. [Crossref] [PubMed]
- Brackmann F, Kehrer C, Kustermann W, et al. Rare Variant of GM2 Gangliosidosis through Activator-Protein Deficiency. Neuropediatrics 2017;48:127-30. [Crossref] [PubMed]
- Ganne B, Dauriat B, Richard L, et al. GM2 gangliosidosis AB variant: first case of late onset and review of the literature. Neurol Sci 2022;43:6517-27. [Crossref] [PubMed]
- İnci A, Cengiz Ergin FB, Biberoğlu G, et al. Two patients from Turkey with a novel variant in the GM2A gene and review of the literature. J Pediatr Endocrinol Metab 2021;34:805-12. [Crossref] [PubMed]
- Encarnação M, Coutinho MF, Silva L, et al. Assessing Lysosomal Disorders in the NGS Era: Identification of Novel Rare Variants. Int J Mol Sci 2020;21:6355. [Crossref] [PubMed]
- Kolodny E, Sathe S, Zeng BJ, et al. 55. A novel GM2-activator deficiency mutation as a cause of AB variant GM2-Gangliosidosis. Molecular Genetics and Metabolism 2008;93:27-8. [Crossref]
- Kochumon SP, Yesodharan D, Vinayan K, et al. GM2 activator protein deficiency, mimic of Tay-Sachs disease. International Journal of Epilepsy 2017;4:184-7. [Crossref]
- O'Brien JS, Kishimoto Y. Saposin proteins: structure, function, and role in human lysosomal storage disorders. FASEB J 1991;5:301-8. [Crossref] [PubMed]
- Rorman EG, Scheinker V, Grabowski GA. Structure and evolution of the human prosaposin chromosomal gene. Genomics 1992;13:312-8. [Crossref] [PubMed]
- Yuan L, Morales CR. A stretch of 17 amino acids in the prosaposin C terminus is critical for its binding to sortilin and targeting to lysosomes. J Histochem Cytochem 2010;58:287-300. [Crossref] [PubMed]
- Morimoto S, Martin BM, Yamamoto Y, et al. Saposin A: second cerebrosidase activator protein. Proc Natl Acad Sci U S A 1989;86:3389-93. [Crossref] [PubMed]
- Suzuki K, Suzuki Y. Globoid cell leucodystrophy (Krabbe's disease): deficiency of galactocerebroside beta-galactosidase. Proc Natl Acad Sci U S A 1970;66:302-9. [Crossref] [PubMed]
- Spiegel R, Bach G, Sury V, et al. A mutation in the saposin A coding region of the prosaposin gene in an infant presenting as Krabbe disease: first report of saposin A deficiency in humans. Mol Genet Metab 2005;84:160-6. [Crossref] [PubMed]
- Kose M, Demir SA, Akinci G, et al. The Second Case of Saposin A Deficiency and Altered Autophagy. JIMD Rep 2019;44:43-54. [Crossref] [PubMed]
- Calderwood L, Wenger DA, Matern D, et al. Rare Saposin A deficiency: Novel variant and psychosine analysis. Mol Genet Metab 2020;129:161-4. [Crossref] [PubMed]
- Chang MH, Bindloss CA, Grabowski GA, et al. Saposins A, B, C, and D in plasma of patients with lysosomal storage disorders. Clin Chem 2000;46:167-74. [Crossref] [PubMed]
- Eto Y, Suzuki K. Brain sphingoglycolipids in Krabbe's globoid cell leucodystrophy. J Neurochem 1971;18:503-11. [Crossref] [PubMed]
- Suzuki K. Renal cerebroside in globoid cell leukodystrophy (Krabbe's disease). Lipids 1971;6:433-6. [Crossref] [PubMed]
- Dawson G, Matalon R, Dorfman A. Glycosphingolipids in cultured human skin fibroblasts. II. Characterization and metabolism in fibroblasts from patients with inborn errors of glycosphingolipid and mucopolysaccharide metabolism. J Biol Chem 1972;247:5951-8. [Crossref] [PubMed]
- Kaye EM, Ullman MD, Kolodny EH, et al. Possible use of CSF glycosphingolipids for the diagnosis and therapeutic monitoring of lysosomal storage diseases. Neurology 1992;42:2290-4. [Crossref] [PubMed]
- Dawson G. Hepatic galactosylceramide in globoid cell leukodystrophy (Krabbe's disease). Lipids 1973;8:154-6. [Crossref] [PubMed]
- Svennerholm L, Vanier MT, Månsson JE. Krabbe disease: a galactosylsphingosine (psychosine) lipidosis. J Lipid Res 1980;21:53-64. [Crossref] [PubMed]
- Taketomi T, Hara A, Kutsukake Y, et al. New approach for characterization of lysosulfatide by TLC, fast atom bombardment mass spectrometry and NMR spectroscopy. J Biochem 1990;107:680-4. [Crossref] [PubMed]
- Igisu H, Suzuki K. Analysis of galactosylsphingosine (psychosine) in the brain. J Lipid Res 1984;25:1000-6. [Crossref] [PubMed]
- Nozawa M, Iwamoto T, Tokoro T, et al. Novel procedure for measuring psychosine derivatives by an HPLC method. J Neurochem 1992;59:607-9. [Crossref] [PubMed]
- Whitfield PD, Sharp PC, Taylor R, et al. Quantification of galactosylsphingosine in the twitcher mouse using electrospray ionization-tandem mass spectrometry. J Lipid Res 2001;42:2092-5. [Crossref] [PubMed]
- Esch SW, Williams TD, Biswas S, et al. Sphingolipid profile in the CNS of the twitcher (globoid cell leukodystrophy) mouse: a lipidomics approach. Cell Mol Biol (Noisy-le-grand) 2003;49:779-87. [PubMed]
- Zanfini A, Dreassi E, Berardi A, et al. Quantification of psychosine in the serum of twitcher mouse by LC-ESI-tandem-MS analysis. J Pharm Biomed Anal 2013;80:44-9. [Crossref] [PubMed]
- Chuang WL, Pacheco J, Zhang XK, et al. Determination of psychosine concentration in dried blood spots from newborns that were identified via newborn screening to be at risk for Krabbe disease. Clin Chim Acta 2013;419:73-6. [Crossref] [PubMed]
- Turgeon CT, Orsini JJ, Sanders KA, et al. Measurement of psychosine in dried blood spots--a possible improvement to newborn screening programs for Krabbe disease. J Inherit Metab Dis 2015;38:923-9. [Crossref] [PubMed]
- Escolar ML, Kiely BT, Shawgo E, et al. Psychosine, a marker of Krabbe phenotype and treatment effect. Mol Genet Metab 2017;121:271-8. [Crossref] [PubMed]
- Guenzel AJ, Turgeon CT, Nickander KK, et al. The critical role of psychosine in screening, diagnosis, and monitoring of Krabbe disease. Genet Med 2020;22:1108-18. [Crossref] [PubMed]
- Kwon JM, Matern D, Kurtzberg J, et al. Consensus guidelines for newborn screening, diagnosis and treatment of infantile Krabbe disease. Orphanet J Rare Dis 2018;13:30. [Crossref] [PubMed]
- Beasley J, McCaw P, Zhang H, et al. Combined analysis of plasma or serum glucosylsphingosine and globotriaosylsphingosine by UPLC-MS/MS. Clin Chim Acta 2020;511:132-7. [Crossref] [PubMed]
- Matsumoto SI, Sato S, Otake K, et al. Highly-sensitive simultaneous quantitation of glucosylsphingosine and galactosylsphingosine in human cerebrospinal fluid by liquid chromatography/tandem mass spectrometry. J Pharm Biomed Anal 2022;217:114852. [Crossref] [PubMed]
- Mehl E, Jatzkewitz H. A cerebrosidesulfatase from swine kidney. Hoppe Seylers Z Physiol Chem 1964;339:260-76. [Crossref] [PubMed]
- Inui K, Wenger DA. Biochemical, immunological, and structural studies on a sphingolipid activator protein (SAP-1). Arch Biochem Biophys 1984;233:556-64. [Crossref] [PubMed]
- Gärtner S, Conzelmann E, Sandhoff K. Activator protein for the degradation of globotriaosylceramide by human alpha-galactosidase. J Biol Chem 1983;258:12378-85. [Crossref] [PubMed]
- Vogel A, Fürst W, Abo-Hashish MA, et al. Identity of the activator proteins for the enzymatic hydrolysis of sulfatide, ganglioside GM1, and globotriaosylceramide. Arch Biochem Biophys 1987;259:627-38. [Crossref] [PubMed]
- Austin J, McAfee D, Armstrong D, et al. Abnormal sulphatase activities in two human diseases (metachromatic leucodystrophy and gargoylism). Biochem J 1964;93:15C-7C. [Crossref] [PubMed]
- Kint JA. Fabry's disease: alpha-galactosidase deficiency. Science 1970;167:1268-9. [Crossref] [PubMed]
- Okada S, O'Brien JS. Generalized gangliosidosis: beta-galactosidase deficiency. Science 1968;160:1002-4. [Crossref] [PubMed]
- Shapiro LJ, Aleck KA, Kaback MM, et al. Metachromatic leukodystrophy without arylsulfatase A deficiency. Pediatr Res 1979;13:1179-81. [Crossref] [PubMed]
- Inui K, Emmett M, Wenger DA. Immunological evidence for deficiency in an activator protein for sulfatide sulfatase in a variant form of metachromatic leukodystrophy. Proc Natl Acad Sci U S A 1983;80:3074-7. [Crossref] [PubMed]
- Stevens RL, Fluharty AL, Kihara H, et al. Cerebroside sulfatase activator deficiency induced metachromatic leukodystrophy. Am J Hum Genet 1981;33:900-6. [PubMed]
- Fujibayashi S, Inui K, Wenger DA. Activator protein-deficient metachromatic leukodystrophy: diagnosis in leukocytes using immunologic methods. J Pediatr 1984;104:739-42. [Crossref] [PubMed]
- Fischer G, Jatzkewitz H. The activator of cerebroside sulphatase. Purification from human liver and identification as a protein. Hoppe Seylers Z Physiol Chem 1975;356:605-13. [Crossref] [PubMed]
- Norris AJ, Whitelegge JP, Yaghoubian A, et al. A novel mass spectrometric assay for the cerebroside sulfate activator protein (saposin B) and arylsulfatase A. J Lipid Res 2005;46:2254-64. [Crossref] [PubMed]
- HAGBERG B. SVENNERHOLM L. Metachromatic leucodystrophy-a generalized lipidosis. Determination of sulfatides in urine, blood plasma and cerebrospinal fluid. Acta Paediatr (Stockh) 1960;49:690-4. [Crossref] [PubMed]
- Pollet S, Ermidou S, Le Saux F, et al. Microanalysis of brain lipids: multiple two-dimensional thin-layer chromatography. J Lipid Res 1978;19:916-21. [Crossref] [PubMed]
- Kean EL. Rapid, sensitive spectrophotometric method for quantitative determination of sulfatides. J Lipid Res 1968;9:319-27. [Crossref] [PubMed]
- Strasberg PM, Warren I, Skomorowski MA, et al. HPLC analysis of urinary sulfatide: an aid in the diagnosis of metachromatic leukodystrophy. Clin Biochem 1985;18:92-7. [Crossref] [PubMed]
- Berná L, Asfaw B, Conzelmann E, et al. Determination of urinary sulfatides and other lipids by combination of reversed-phase and thin-layer chromatographies. Anal Biochem 1999;269:304-11. [Crossref] [PubMed]
- Hsu FF, Bohrer A, Turk J. Electrospray ionization tandem mass spectrometric analysis of sulfatide. Determination of fragmentation patterns and characterization of molecular species expressed in brain and in pancreatic islets. Biochim Biophys Acta 1998;1392:202-16. [Crossref] [PubMed]
- Whitfield PD, Sharp PC, Johnson DW, et al. Characterization of urinary sulfatides in metachromatic leukodystrophy using electrospray ionization-tandem mass spectrometry. Mol Genet Metab 2001;73:30-7. [Crossref] [PubMed]
- Cui Y, Colsch B, Afonso C, et al. Synthetic sulfogalactosylceramide (sulfatide) and its use for the mass spectrometric quantitative urinary determination in metachromatic leukodystrophies. Glycoconj J 2008;25:147-55. [Crossref] [PubMed]
- Kuchař L, Asfaw B, Poupětová H, et al. Direct tandem mass spectrometric profiling of sulfatides in dry urinary samples for screening of metachromatic leukodystrophy. Clin Chim Acta 2013;425:153-9. [Crossref] [PubMed]
- Han M, Jun SH, Song SH, et al. Ultra-performance liquid chromatography/tandem mass spectrometry for determination of sulfatides in dried blood spots from patients with metachromatic leukodystrophy. Rapid Commun Mass Spectrom 2014;28:587-94. [Crossref] [PubMed]
- Barcenas M, Suhr TR, Scott CR, et al. Quantification of sulfatides in dried blood and urine spots from metachromatic leukodystrophy patients by liquid chromatography/electrospray tandem mass spectrometry. Clin Chim Acta 2014;433:39-43. [Crossref] [PubMed]
- Toda K, Kobayashi T, Goto I, et al. Accumulation of lysosulfatide (sulfogalactosylsphingosine) in tissues of a boy with metachromatic leukodystrophy. Biochem Biophys Res Commun 1989;159:605-11. [Crossref] [PubMed]
- Kyogashima M, Tamiya-Koizumi K, Ehara T, et al. Rapid demonstration of diversity of sulfatide molecular species from biological materials by MALDI-TOF MS. Glycobiology 2006;16:719-28. [Crossref] [PubMed]
- Sugiyama E, Hara A, Uemura K. A quantitative analysis of serum sulfatide by matrix-assisted laser desorption ionization time-of-flight mass spectrometry with delayed ion extraction. Anal Biochem 1999;274:90-7. [Crossref] [PubMed]
- Taketomi T, Hara A, Uemura K, et al. A microwave-mediated saponification of galactosylceramide and galactosylceramide I3-sulfate and identification of their lyso-compounds by delayed extraction matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Biochem Biophys Res Commun 1996;224:462-7. [Crossref] [PubMed]
- Mirzaian M, Kramer G, Poorthuis BJ. Quantification of sulfatides and lysosulfatides in tissues and body fluids by liquid chromatography-tandem mass spectrometry. J Lipid Res 2015;56:936-43. [Crossref] [PubMed]
- Cao KH, Zhang G, Jacobsen L, et al. LC-MS/MS assays to quantify sulfatides and lysosulfatide in cerebrospinal fluid of metachromatic leukodystrophy patients. Bioanalysis 2020;12:1621-33. [Crossref] [PubMed]
- Carnicer-Cáceres C, Arranz-Amo JA, Cea-Arestin C, et al. Biomarkers in Fabry Disease. Implications for Clinical Diagnosis and Follow-up. J Clin Med 2021;10:1664. [Crossref] [PubMed]
- Ogawa K, Fujiwara Y, Sugamata K, et al. Thin-layer chromatography of neutral glycosphingolipids: an improved simple method for the resolution of GlcCer, GalCer, LacCer and Ga2Cer. J Chromatogr 1988;426:188-93. [Crossref] [PubMed]
- Ullman MD, McCluer RH. Quantitative analysis of plasma neutral glycosphingolipids by high performance liquid chromatography of their perbenzoyl derivatives. J Lipid Res 1977;18:371-8. [Crossref] [PubMed]
- Vance DE, Sweeley CC. Quantitative determination of the neutral glycosyl ceramides in human blood. J Lipid Res 1967;8:621-30. [Crossref] [PubMed]
- Zeidner KM, Desnick RJ, Ioannou YA. Quantitative determination of globotriaosylceramide by immunodetection of glycolipid-bound recombinant verotoxin B subunit. Anal Biochem 1999;267:104-13. [Crossref] [PubMed]
- Boscaro F, Pieraccini G, la Marca G, et al. Rapid quantitation of globotriaosylceramide in human plasma and urine: a potential application for monitoring enzyme replacement therapy in Anderson-Fabry disease. Rapid Commun Mass Spectrom 2002;16:1507-14. [Crossref] [PubMed]
- Auray-Blais C, Cyr D, Mills K, et al. Development of a filter paper method potentially applicable to mass and high-risk urinary screenings for Fabry disease. J Inherit Metab Dis 2007;30:106. [Crossref] [PubMed]
- Mills K, Johnson A, Winchester B. Synthesis of novel internal standards for the quantitative determination of plasma ceramide trihexoside in Fabry disease by tandem mass spectrometry. FEBS Lett 2002;515:171-6. [Crossref] [PubMed]
- Young E, Mills K, Morris P, et al. Is globotriaosylceramide a useful biomarker in Fabry disease? Acta Paediatr Suppl 2005;94:51-4; discussion 37-8. [Crossref] [PubMed]
- Kuchar L, Ledvinová J, Hrebícek M, et al. Prosaposin deficiency and saposin B deficiency (activator-deficient metachromatic leukodystrophy): report on two patients detected by analysis of urinary sphingolipids and carrying novel PSAP gene mutations. Am J Med Genet A 2009;149A:613-21. [Crossref] [PubMed]
- Ferraz MJ, Marques AR, Appelman MD, et al. Lysosomal glycosphingolipid catabolism by acid ceramidase: formation of glycosphingoid bases during deficiency of glycosidases. FEBS Lett 2016;590:716-25. [Crossref] [PubMed]
- Togawa T, Kodama T, Suzuki T, et al. Plasma globotriaosylsphingosine as a biomarker of Fabry disease. Mol Genet Metab 2010;100:257-61. [Crossref] [PubMed]
- Auray-Blais C, Ntwari A, Clarke JT, et al. How well does urinary lyso-Gb3 function as a biomarker in Fabry disease? Clin Chim Acta 2010;411:1906-14. [Crossref] [PubMed]
- Gold H, Mirzaian M, Dekker N, et al. Quantification of globotriaosylsphingosine in plasma and urine of fabry patients by stable isotope ultraperformance liquid chromatography-tandem mass spectrometry. Clin Chem 2013;59:547-56. [Crossref] [PubMed]
- Lavoie P, Boutin M, Auray-Blais C. Multiplex analysis of novel urinary lyso-Gb3-related biomarkers for Fabry disease by tandem mass spectrometry. Anal Chem 2013;85:1743-52. [Crossref] [PubMed]
- Auray-Blais C, Boutin M. Novel gb(3) isoforms detected in urine of fabry disease patients: a metabolomic study. Curr Med Chem 2012;19:3241-52. [Crossref] [PubMed]
- Abaoui M, Boutin M, Lavoie P, et al. Tandem mass spectrometry multiplex analysis of methylated and non-methylated urinary Gb3 isoforms in Fabry disease patients. Clin Chim Acta 2016;452:191-8. [Crossref] [PubMed]
- Paschke E, Kresse H. Morquio disease, type B: activation of GM1-beta-galactosidase by GM1-activator protein. Biochem Biophys Res Commun 1982;109:568-75. [Crossref] [PubMed]
- Hiraiwa M, Martin BM, Kishimoto Y, et al. Lysosomal proteolysis of prosaposin, the precursor of saposins (sphingolipid activator proteins): its mechanism and inhibition by ganglioside. Arch Biochem Biophys 1997;341:17-24. [Crossref] [PubMed]
- Callahan JW. Molecular basis of GM1 gangliosidosis and Morquio disease, type B. Structure-function studies of lysosomal beta-galactosidase and the non-lysosomal beta-galactosidase-like protein. Biochim Biophys Acta 1999;1455:85-103. [Crossref] [PubMed]
- Schmid B, Paton BC, Sandhoff K, et al. Metabolism of GM1 ganglioside in cultured skin fibroblasts: anomalies in gangliosidoses, sialidoses, and sphingolipid activator protein (SAP, saposin) 1 and prosaposin deficient disorders. Hum Genet 1992;89:513-8. [Crossref] [PubMed]
- Sheth J, Nair A, Bhavsar R, et al. Late infantile and adult-onset metachromatic leukodystrophy due to novel missense variants in the PSAP gene: Case report from India. JIMD Rep 2023;64:265-73. [Crossref] [PubMed]
- Alfadhel M, Almuqbil M, Al Mutairi F, et al. The Leukodystrophy Spectrum in Saudi Arabia: Epidemiological, Clinical, Radiological, and Genetic Data. Front Pediatr 2021;9:633385. [Crossref] [PubMed]
- Al-Hassnan ZN, Al Dhalaan H, Patay Z, et al. Sphingolipid activator protein B deficiency: report of 9 Saudi patients and review of the literature. J Child Neurol 2009;24:1513-9. [Crossref] [PubMed]
- Ho MW, O'Brien JS. Gaucher's disease: deficiency of 'acid' -glucosidase and reconstitution of enzyme activity in vitro. Proc Natl Acad Sci U S A 1971;68:2810-3. [Crossref] [PubMed]
- Poulos A, Ranieri E, Shankaran P, et al. Studies on the activation of the enzymatic hydrolysis of sphingomyelin liposomes. Biochim Biophys Acta 1984;793:141-8. [Crossref] [PubMed]
- Wenger DA, Sattler M, Roth S. A protein activator of galactosylceramide beta-galactosidase. Biochim Biophys Acta 1982;712:639-49. [Crossref] [PubMed]
- Vaccaro AM, Muscillo M, Salvioli R, et al. The binding of glucosylceramidase to glucosylceramide is promoted by its activator protein. FEBS Lett 1987;216:190-4. [Crossref] [PubMed]
- Christomanou H, Aignesberger A, Linke RP. Immunochemical characterization of two activator proteins stimulating enzymic sphingomyelin degradation in vitro. Absence of one of them in a human Gaucher disease variant. Biol Chem Hoppe Seyler 1986;367:879-90. [Crossref] [PubMed]
- Tylki-Szymańska A, Czartoryska B, Vanier MT, et al. Non-neuronopathic Gaucher disease due to saposin C deficiency. Clin Genet 2007;72:538-42. [Crossref] [PubMed]
- Christomanou H, Chabás A, Pámpols T, et al. Activator protein deficient Gaucher's disease. A second patient with the newly identified lipid storage disorder. Klin Wochenschr 1989;67:999-1003. [Crossref] [PubMed]
- Meikle PJ, Ranieri E, Simonsen H, et al. Newborn screening for lysosomal storage disorders: clinical evaluation of a two-tier strategy. Pediatrics 2004;114:909-16. [Crossref] [PubMed]
- Yoneshige A, Muto M, Watanabe T, et al. The effects of chemically synthesized saposin C on glucosylceramide-β-glucosidase. Clin Biochem 2015;48:1177-80. [Crossref] [PubMed]
- Aerts JM, Kallemeijn WW, Wegdam W, et al. Biomarkers in the diagnosis of lysosomal storage disorders: proteins, lipids, and inhibodies. J Inherit Metab Dis 2011;34:605-19. [Crossref] [PubMed]
- Dekker N, van Dussen L, Hollak CE, et al. Elevated plasma glucosylsphingosine in Gaucher disease: relation to phenotype, storage cell markers, and therapeutic response. Blood 2011;118:e118-27. [Crossref] [PubMed]
- Mohamed FE, Ali A, Al-Tenaiji A, et al. A Type 3 Gaucher-Like Disease Due To Saposin C Deficiency in Two Emirati Families Caused by a Novel Splice Site Variant in the PSAP Gene. J Mol Neurosci 2022;72:1322-33. [Crossref] [PubMed]
- Vance DE, Krivit W, Sweeley CC. Concentrations of glycosyl ceramides in plasma and red cells in Fabry's disease, a glycolipid lipidosis. J Lipid Res 1969;10:188-92. [Crossref] [PubMed]
- Evans JE, McCluer RH. High pressure liquid chromatography of neutral glycosphingolipids. Biochim Biophys Acta 1972;270:565-9. [Crossref] [PubMed]
- Strasberg PM, Warren I, Skomorowski MA, et al. HPLC analysis of neutral glycolipids: an aid in the diagnosis of lysosomal storage disease. Clin Chim Acta 1983;132:29-41. [Crossref] [PubMed]
- Samuelsson B, Samuelsson K. Gas-liquid chromatographic separation of ceramides as di-O-trimethylsilyl ether derivatives. Biochim Biophys Acta 1968;164:421-3. [Crossref] [PubMed]
- Murata T, Ariga T, Oshima M, et al. Characterization of trimethylsilyl derivatives of cerebrosides by direct inlet-chemical ionization mass spectrometry. J Lipid Res 1978;19:370-4. [Crossref] [PubMed]
- HILLBORG PO. SVENNERHOLM L. Blood level of cerebrosides in Gaucher's disease. Acta Paediatr (Stockh) 1960;49:707-10. [Crossref] [PubMed]
- Whitfield PD, Nelson P, Sharp PC, et al. Correlation among genotype, phenotype, and biochemical markers in Gaucher disease: implications for the prediction of disease severity. Mol Genet Metab 2002;75:46-55. [Crossref] [PubMed]
- Raghavan SS, Mumford RA, Kanfer JN. Deficiency of glucosylsphingosine: beta-glucosidase in Gaucher disease. Biochem Biophys Res Commun 1973;54:256-63. [Crossref] [PubMed]
- Raghavan SS, Mumford RA, Kanfer JN. Isolation and characterization of glucosylsphingosine from Gaucher's spleen. J Lipid Res 1974;15:484-90. [Crossref] [PubMed]
- Orvisky E, Sidransky E, McKinney CE, et al. Glucosylsphingosine accumulation in mice and patients with type 2 Gaucher disease begins early in gestation. Pediatr Res 2000;48:233-7. [Crossref] [PubMed]
- Orvisky E, Park JK, LaMarca ME, et al. Glucosylsphingosine accumulation in tissues from patients with Gaucher disease: correlation with phenotype and genotype. Mol Genet Metab 2002;76:262-70. [Crossref] [PubMed]
- Oshima M. Identification of glucosyl sphingosine from Gaucher's spleen by gas chromatography-electron impact and GC-chemical ionization mass spectrometry. J Biochem 1976;80:53-9. [Crossref] [PubMed]
- Kobayashi T, Mitsuo K, Goto I. Free sphingoid bases in normal murine tissues. Eur J Biochem 1988;172:747-52. [Crossref] [PubMed]
- Rolfs A, Giese AK, Grittner U, et al. Glucosylsphingosine is a highly sensitive and specific biomarker for primary diagnostic and follow-up monitoring in Gaucher disease in a non-Jewish, Caucasian cohort of Gaucher disease patients. PLoS One 2013;8:e79732. [Crossref] [PubMed]
- Mirzaian M, Wisse P, Ferraz MJ, et al. Mass spectrometric quantification of glucosylsphingosine in plasma and urine of type 1 Gaucher patients using an isotope standard. Blood Cells Mol Dis 2015;54:307-14. [Crossref] [PubMed]
- Zhang W, Oehrle M, Prada CE, et al. A convenient approach to facilitate monitoring Gaucher disease progression and therapeutic response. Analyst 2017;142:3380-7. [Crossref] [PubMed]
- Revel-Vilk S, Fuller M, Zimran A. Value of Glucosylsphingosine (Lyso-Gb1) as a Biomarker in Gaucher Disease: A Systematic Literature Review. Int J Mol Sci 2020;21:7159. [Crossref] [PubMed]
- Menkovic I, Boutin M, Alayoubi A, et al. Identification of a Reliable Biomarker Profile for the Diagnosis of Gaucher Disease Type 1 Patients Using a Mass Spectrometry-Based Metabolomic Approach. Int J Mol Sci 2020;21:7869. [Crossref] [PubMed]
- Menkovic I, Boutin M, Alayoubi A, et al. Quantitation of a plasma biomarker profile for the early detection of Gaucher disease type 1 patients. Bioanalysis 2022;14:223-40. [Crossref] [PubMed]
- Menkovic I, Boutin M, Alayoubi A, et al. Metabolomic Study Using Time-of-Flight Mass Spectrometry Reveals Novel Urinary Biomarkers for Gaucher Disease Type 1. J Proteome Res 2022;21:1321-9. [Crossref] [PubMed]
- Menkovic I, Boutin M, Alayoubi A, et al. Quantitation of a Urinary Profile of Biomarkers in Gaucher Disease Type 1 Patients Using Tandem Mass Spectrometry. Diagnostics (Basel) 2022;12:1414. [Crossref] [PubMed]
- Liaqat K, Hussain S, Acharya A, et al. Phenotype Expansion for Atypical Gaucher Disease Due to Homozygous Missense PSAP Variant in a Large Consanguineous Pakistani Family. Genes (Basel) 2022;13:662. [Crossref] [PubMed]
- Radha Rama Devi A, Kadali S, Radhika A, et al. Acute Gaucher Disease-Like Condition in an Indian Infant with a Novel Biallelic Mutation in the Prosaposin Gene. J Pediatr Genet 2019;8:81-5. [Crossref] [PubMed]
- Amsallem D, Rodriguez D, Vanier MT. Third case of Gaucher disease with sap-C deficiency and evaluation of twelve months’ therapy by miglustat. J Inherit Metab Dis 2005;28:152-5.
- Morimoto S, Martin BM, Kishimoto Y, et al. Saposin D: a sphingomyelinase activator. Biochem Biophys Res Commun 1988;156:403-10. [Crossref] [PubMed]
- Azuma N, O'Brien JS, Moser HW, et al. Stimulation of acid ceramidase activity by saposin D. Arch Biochem Biophys 1994;311:354-7. [Crossref] [PubMed]
- Matsuda J, Kido M, Tadano-Aritomi K, et al. Mutation in saposin D domain of sphingolipid activator protein gene causes urinary system defects and cerebellar Purkinje cell degeneration with accumulation of hydroxy fatty acid-containing ceramide in mouse. Hum Mol Genet 2004;13:2709-23. [Crossref] [PubMed]
- Sun Y, Witte DP, Zamzow M, et al. Combined saposin C and D deficiencies in mice lead to a neuronopathic phenotype, glucosylceramide and alpha-hydroxy ceramide accumulation, and altered prosaposin trafficking. Hum Mol Genet 2007;16:957-71. [Crossref] [PubMed]
- Oji Y, Hatano T, Ueno SI, et al. Variants in saposin D domain of prosaposin gene linked to Parkinson's disease. Brain 2020;143:1190-205. [Crossref] [PubMed]
- Facchi D, Rimoldi V, Straniero L, et al. Saposin D variants are not a common cause of familial Parkinson's disease among Italians. Brain 2020;143:e71. [Crossref] [PubMed]
- Sosero YL, Bandres-Ciga S, Hassin-Baer S, et al. Lack of evidence for genetic association of saposins A, B, C and D with Parkinson's disease. Brain 2020;143:e72. [Crossref] [PubMed]
- Harzer K, Paton BC, Poulos A, et al. Sphingolipid activator protein deficiency in a 16-week-old atypical Gaucher disease patient and his fetal sibling: biochemical signs of combined sphingolipidoses. Eur J Pediatr 1989;149:31-9. [Crossref] [PubMed]
- Schnabel D, Schröder M, Fürst W, et al. Simultaneous deficiency of sphingolipid activator proteins 1 and 2 is caused by a mutation in the initiation codon of their common gene. J Biol Chem 1992;267:3312-5. [Crossref] [PubMed]
- Paton BC, Schmid B, Kustermann-Kuhn B, et al. Additional biochemical findings in a patient and fetal sibling with a genetic defect in the sphingolipid activator protein (SAP) precursor, prosaposin. Evidence for a deficiency in SAP-1 and for a normal lysosomal neuraminidase. Biochem J 1992;285:481-8. [Crossref] [PubMed]
- Bradová V, Smíd F, Ulrich-Bott B, et al. Prosaposin deficiency: further characterization of the sphingolipid activator protein-deficient sibs. Multiple glycolipid elevations (including lactosylceramidosis), partial enzyme deficiencies and ultrastructure of the skin in this generalized sphingolipid storage disease. Hum Genet 1993;92:143-52. [Crossref] [PubMed]
- Elleder M, Jerábková M, Befekadu A, et al. Prosaposin deficiency -- a rarely diagnosed, rapidly progressing, neonatal neurovisceral lipid storage disease. Report of a further patient. Neuropediatrics 2005;36:171-80. [Crossref] [PubMed]
- Hulková H, Cervenková M, Ledvinová J, et al. A novel mutation in the coding region of the prosaposin gene leads to a complete deficiency of prosaposin and saposins, and is associated with a complex sphingolipidosis dominated by lactosylceramide accumulation. Hum Mol Genet 2001;10:927-40. [Crossref] [PubMed]
- Bhat V, Thergaonkar RW, Thakur M, et al. Combined saposin deficiency: A rare occurrence. Med J Armed Forces India 2023;79:238-40. [Crossref] [PubMed]
- Motta M, Tatti M, Furlan F, et al. Clinical, biochemical and molecular characterization of prosaposin deficiency. Clin Genet 2016;90:220-9. [Crossref] [PubMed]
- Elleder M, Ledvinová J, Hřebíček M, et al. editors. Book of Abstracts 14th ESGLD Workshop (September 18th-21st, 2003). Podebrady/Prague: Guarant Ltd; 2003.
Cite this article as: Yuan L. Unraveling the diagnostic enigma: laboratory diagnosis of sphingolipid activator protein deficiencies. J Lab Precis Med 2024;9:22.


